Analyzing Preprints: The challenges of working with metadata from bioRxiv
By Mario Malički, Maria Janina Sarol, and Juan Pablo Alperin
This blog post is the third in a four part series documenting the methodological challenges we faced during our project investigating preprint growth and uptake. Stay tuned for our final post, in which we’ll dive into the arXiv metadata.

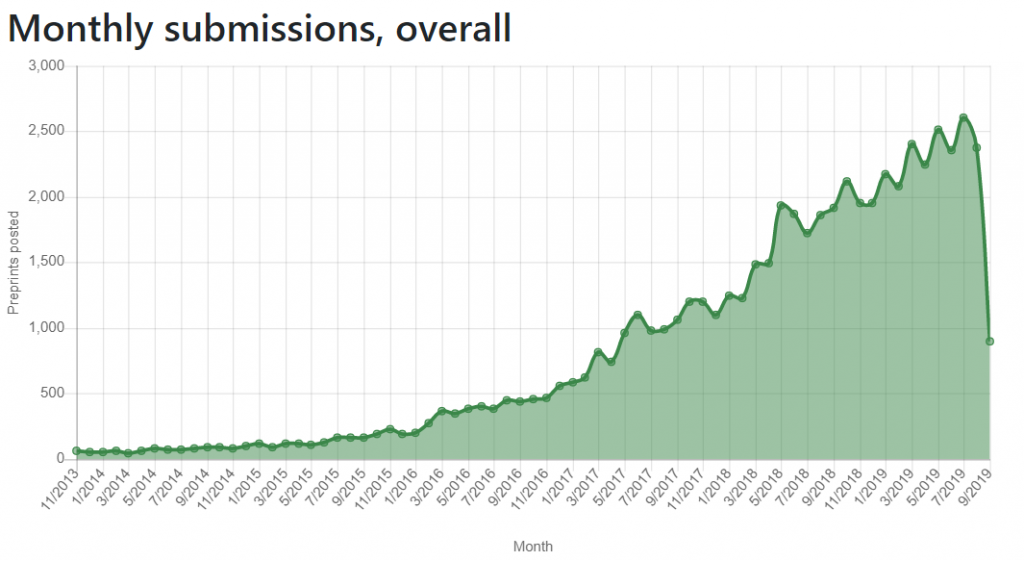
Following our analysis of the challenges working with SHARE and OSF metadata (read about them in our previous blogs here and here), we explored the metadata of bioRxiv—a preprint server focused on biology research. Since its inception in 2013, bioRxiv has received significant attention,1 and became the largest biology-focused preprint server (Figure 1, below). Studies showed that posting a preprint on bioRxiv was associated with more citations and higher Altmetric scores than articles that were not posted as preprints.2-4 Additionally, recent research (itself posted on bioRxiv) has shown slight differences between preprints posted on bioRxiv and postprints published in journals indexed in PubMed, with peer reviewed articles having better reporting of reagents (i.e., drug suppliers and antibody validation) and experimental animals (i.e., reporting of strain, sex, supplier and randomization), but bioRxiv articles having better reporting of unit-level data, completeness of statistical results, and exact p-values.5
Unlike OSF servers we previously analysed, bioRxiv only accepts unpublished/unrefereed manuscripts that have not been peer reviewed or accepted for publication in a journal (Figure 1, below). Furthermore, bioRxiv specifically states it does not accept theses, dissertations, or protocols. It accepts only preprints written in English and uploaded as either Word documents or PDFs. Moreover, all uploaded preprints, including their updated versions, undergo a basic screening process for content that is offensive, non-scientific, or that might pose a health or biosecurity risk, and undergo iThenthicate plagiarism check and duplicate search (information provided by bioRxiv). To encourage use of preprints, bioRxiv allows users to directly transmit uploaded documents and their metadata to a number of journals, and journals to deposit received manuscripts as preprints to bioRxiv.
Despite not yet releasing their own public API, bioRxiv metadata can be retrieved from Crossref and through the Rxivist site. BioRxiv deposits metadata to Crossref as part of the official scholarly record; while the metadata available from Rxivist is scraped from the bioRxiv website.6
For our analysis, we downloaded metadata from June 18 to 20, 2019 for all posted content in Crossref—that is, all “preprints, eprints, working papers, reports, dissertations, and many other types of content that has been posted but not formally published”—and filtered it for records from bioRxiv. Guided by a description of Crossref metadata fields, we then set out to explore the same basic metadata fields as we did for SHARE and OSF. Here’s what we found:
1) Records
Our final database contained 52,531 preprints. Although bioRxiv users have to categorize their preprints into one of three categories (New Results, Confirmatory Results, or Contradictory Results), this information is only available through the biorXiv search engine—and not in the Crossref or Rxivist metadata. On August 28, 405 (0.7%) preprints were listed as Contradictory and 1,090 (1.9%) as Confirmatory; all the rest were classified as New Results.
Although records can be removed in cases of discovered plagiarism or breach of relevant ethical standards (see the bioRxiv Submission Guide for details), there was no metadata field for withdrawn status in either Crossref or Rxivist. However, the bioRxiv’s search engine includes a filter option that on August 28 listed 49 preprints as withdrawn. Unlike for published papers, for which the Committee on Publication Ethics recommends keeping the full text of the record available after indicating its retracted or withdrawn status, some uploaded documents of withdrawn preprints in bioRxiv, are removed from the server, and their abstracts replaced with removal reason statements. This is similar to the practice we found in OSF.
There is also no metadata information available for authors of the removal statements, although, in some cases, the statements themselves contain information about the author (e.g., as in example one, a preprint removed by the bioRxiv moderators, or example two, a preprint removed at the request of the authors). However, in several cases the reason for removal is not clear (e.g., example three, Figure 2 below).

Using Crossref Metadata, we also identified 10 duplicate cases, where the title and author(s) of one preprint was an exact match to that of one or more other preprints (e.g., example one, preprints one and two are identical; example two, preprints one, two, and three are different versions of the same preprint). It is worth nothing that bioRxiv has recently implemented a duplicate search.
2) Subjects
On upload, bioRxiv users have to assign their preprint into one of 25 subject areas. Previously users could assign preprints to two additional subject areas – Clinical Trials and Epidemiology – until content in these areas were redirected to medRxiv, a preprint server for clinical sciences launched in June. Taking these two discontinued subject areas into account, we originally expected the bioRxiv metadata to include 27 subject area classifications. Instead, the Crossref metadata listed 29 areas, including two areas not listed on the bioRxiv website: Scientific Communication (n=16) and Pharmacology (n=26), which existed alongside listed Scientific Communication and Education (n=322), and Pharmacology and Toxicology (n=400) areas. (A recent update of bioRxiv metadata has corrected this.)
There were only eight records (0.02%) which did not have a subject information in the Crossref metadata. Of these eight, only two listed subject information on the bioRxiv website and in Rxivist (i.e., records one and two); the remaining six did not (i.e., records one, two, three, four, five, and six).
3) Dates
There are six date fields in the Crossref metadata for bioRxiv preprints: indexed, posted, issued, accepted, created, and deposited. All six of these fields were populated for each of the bioRxiv preprints in our database.
Crossref defines these fields as follows:
- Date indexed: when the metadata was most recently processed by Crossref;
- Date posted: when the content was made available online;
- Date issued: the earliest known publication date. This was equal to date posted in all the cases we studied;
- Date accepted: the date on which a work was accepted for publication. For 37,730 (72%) records, this was equal to the date posted/issued, for 13,517 (26%), it preceded the date posted, and for 1290 (2%) it succeeded it. A description of why this is so, would be a welcome addition to the metadata documentation;
- Date created: when the DOI was first registered. For all but one preprint, Figure 3 below, this date was the same or succeeded the date posted/issued—likely because most DOIs are assigned on the same day or a few days after a preprint is created on the bioRxiv server.
- Date deposited: when the metadata was last updated (by bioRxiv).
The Rxivist metadata contains only one date field, the date when a preprint was first posted. Metadata regarding version numbers or update dates is not available from either Crossref or Rxivist. However, it is possible to discern the version number by looking at the last two characters of the biorxiv_url field in Rxivist (e.g., example one, whose biorxiv_url ends with v7).
4) Contributors
When entering author information during preprint upload at bioRxiv, users are required to fill in each author’s given and family name and institutional affiliation. They can also optionally fill in the author’s middle name, middle name initials, name suffix, home page, and ORCID iD. Alternatively, users can choose to add a Collaborative Group/Consortium as an author, by filling in a free text field. This information, alongside the uploaded document, is then used to create a final pdf version of the preprints (which users have to approve), and from which the final metadata is extracted.
Only three records in the Crossref metadata lacked any author information (specifically, records one, two, and three—this third one has since been corrected). All other records included, at minimum, at least one consortium or family name. We found 24 (0.05%) records with an empty given name field. In some cases this was because a consortium name had been entered as a family name (e.g., example one), in others because only a family name had been entered (e.g., examples two and three). Additionally, we identified 33 records where an author’s full name (given and family name together) exactly matched that of another author on the same record (these errors have since been corrected). For 859 (2%) preprints, we found a Collaborative Group/Consortium as one of the authors. Two of these (records one and two) were exclusively authored by a consortium. Of preprints with consortia authors, 156 (18%) listed more than one Consortium. One preprint (see Figure 4, below), listed the same consortium name twice.

We also identified cases where authors names were entered as consortia (e.g., example three), as well as cases where the consortia field was used to enter “zero”, “No”, “None”, or “N/A” (many of these issues have since been corrected). In a handful of cases, different records used different spellings or acronyms for the same consortia (e.g., the Karolinska Schizophrenia Project (KASP) was also spelled as Karolinska Schizophrenia Project KaSP, KaSP, and KaSP Consortium). All of these issues make it difficult to accurately determine the number of unique consortia listed as co-authors.
In the Crossref metadata, each author is assigned a Sequence field identifying them as either the first or an additional author on a preprint. As expected, in almost all cases, the additional authors appeared in the metadata in the same order as on the bioRxiv webpage. However, in Crossref JSON metadata format consortia always occupy the last spot of the author list (e.g. example one, where a consortia is the middle position of the author byline, but last in the metadata), which makes it impossible to use Crossref JSON metadata format to determine byline order for preprints co-authored with consortia. Crossref XML metadata format and Rxivist metadata retains the correct author order (note: they do not contain a field indicating exact byline position).
For 28,663 (55%) preprints in Crossref metadata, at least one author had an ORCID iD; for 4,177 (8%), all of the authors did. For 13 preprints, two or more authors shared the same ORCID iD (e.g., example one and two).
Lastly, although bioRxiv requires uploaders to add affiliation information for all authors during upload, this information isn’t captured in the Crossref metadata (though it is available in Rxivist).
5) Linking preprints to postprints
As Indicated in our previous blogposts, it is important to identify if a record on a preprint server is actually a preprint. BioRxiv monitors title and authors of records in Crossref and PubMed. On identifying a potential match, they send an email to the corresponding author for confirmation, and then add that information to the preprint metadata. Additionally, users can also manually alert bioRxiv of postprint publications.3
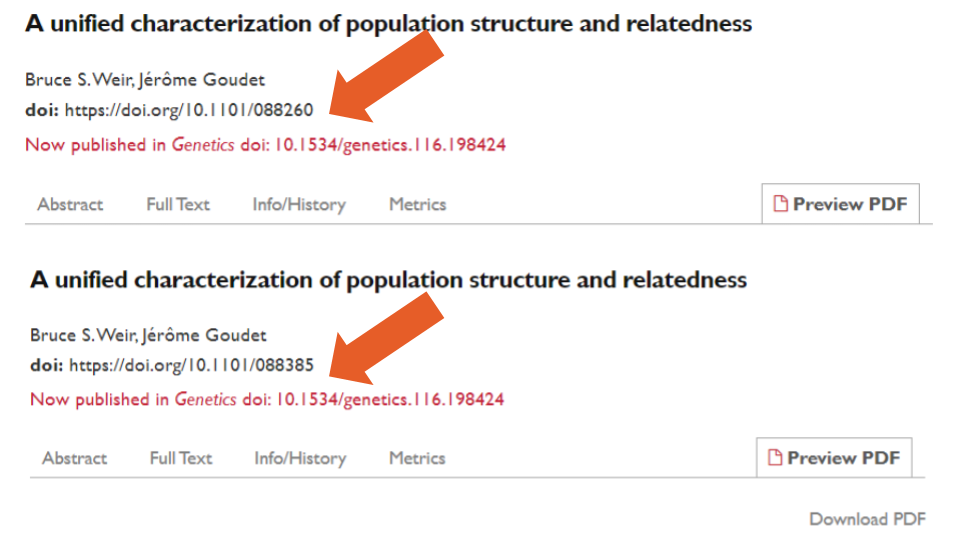
In the Crossref metadata, 20,465 (39%) records had a link to a DOI of their published version. Of those, one record contained its own DOI instead of that of the published postprint (at the time of writing this error had been corrected on the bioRxiv site but not in the metadata deposited to Crossref). We found an additional 29 pairs of preprints that shared an identical postprint DOI with each other. In some cases, this was caused by different versions of the same preprint being uploaded to bioRxiv as separate preprints (e.g., example one: preprints one and two, example two: preprints three and four). In others, the same version had been uploaded twice as two separate preprints (e.g., example three: preprints five and six, Figure 5, below). Additionally, despite accepting only original research preprints, both review papers and published papers have been found among bioRxiv records6-7 (discussion of the preprint community and bioRxiv moderators about these issues can be found here).
Finally, five preprints contained links to reviews and endorsements received on F1000 Prime in Crossref’s relation metadata field (e.g., example one).
Metadata continues to improve
In the two months that have passed since we first downloaded the bioRxiv Crossref metadata and writing of this post, the bioRxiv team has been working and improving the metadata they provide. We hope that they similarly address the errors and omissions we’ve identified above, and enrich the metadata with information about author affiliations, withdrawn records, article categorization, versions, and comments. Overall, we identified fewer issues with bioRxiv metadata than we did for SHARE and OSF. We hope researchers using bioRxiv metadata from either Crossref or Rxivist will report if and how they overcame the challenges we described.
References
- Kaiser J. Are preprints the future of biology? A survival guide for scientists. Science. 2017;397.
- Fu DY, Hughey JJ. Releasing a preprint is associated with more attention and citations. bioRxiv. 2019:699652.
- Fraser N, Momeni F, Mayr P, Peters I. The effect of bioRxiv preprints on citations and altmetrics. bioRxiv. 2019:673665.
- Serghiou S, Ioannidis JP. Altmetric Scores, Citations, and Publication of Studies Posted as Preprints. JAMA: The Journal of the American Medical Association. 2018;319(4):402-4.
- Carneiro CFD, Queiroz VGS, Moulin TC, Carvalho CAM, Haas CB, Rayêe D, et al. Comparing quality of reporting between preprints and peer-reviewed articles in the biomedical literature. bioRxiv. 2019:581892.
- Abdill RJ, Blekhman R. Meta-Research: Tracking the popularity and outcomes of all bioRxiv preprints. eLife. 2019;8:e45133.
- Anaya J. 2018. [cited 2019]. Available from: https://medium.com/@OmnesRes/the-biorxiv-wall-of-shame-aa3d9cfc4cd7.

[…] related metadata. In this post, we draw from our experiences working with metadata from SHARE, OSF, bioRxiv, and arXiv to provide four basic recommendations for those building or managing preprint […]