Analyzing preprints: The challenges of working with metadata from arXiv’s Quantitative Biology section
By Mario Malički, Maria Janina Sarol, and Juan Pablo Alperin
This blog post is the last of a four part series documenting the methodological challenges we faced during our project investigating preprint growth and uptake. Stay tuned for our next post, in which we’ll share general recommendations and hopes for the future of preprint metadata.
Following our analyses of SHARE, OSF, and bioRxiv metadata, (which you can read about here, here, and here), we went on to explore the metadata of arXiv’s Quantitative Biology section.
Started in 1991 as a server for theoretical high-energy physics preprints, arXiv has since expanded to cover many other disciplines and has become the largest repository of preprints in science.1 Its growth has made it an important part of research communication in many fields. Research has shown that usage of arXiv has increased dramatically in the last 10 years in computer science,2-3 as well as in library and information science.4 Research also showed that, across all records and disciplines, 64% of arXiv preprints are eventually published in journals indexed in Web of Science (WoS), and, in turn, journals in WoS are increasingly citing arXiv records.5 arXiv subsection dedicated to Quantitative Biology started in 2003 (Figure 1, below).
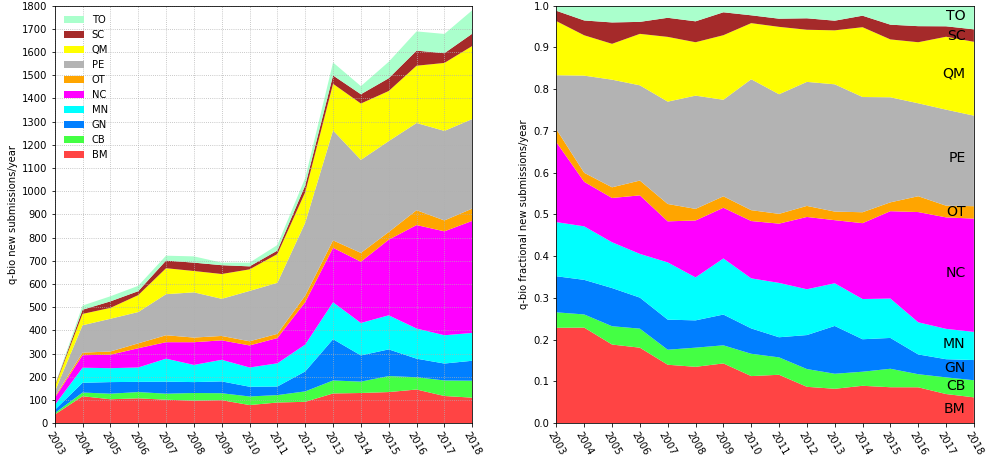
How does q-bio work?
ArXiv differs from other servers we’ve analyzed in some key ways. First, unlike submissions uploaded to OSF and bioRxiv, arXiv preprints are not assigned a DOI, but an arXiv identifier. In addition, although all uploaders on arXiv (whether they are authors or third-party submitters) must assign their submission to a primary section (e.g., q-bio), and they can also subsequently “cross-list” it to other arXiv sections to increase their audience.
On September 3, 2019, we downloaded the metadata for all arXiv records that were listed or cross-listed to q-bio using arXiv’s Open Archives Initiative Protocol for Metadata Harvesting compatible API. Unlike the metadata of SHARE, OSF, bioRxiv, and Rxivist, which is distributed only in JSON, arXiv metadata can be obtained in four XML formats. We based our analyses on arXivRaw — the format that most closely matches arXiv’s internal format. We then explored the same metadata fields we investigated for SHARE, OSF, and bioRxiv (see our previous posts). Additional details of the various metadata fields available can be found on arXiv’s help pages.
1) Records
There were 28,104 records categorized as belonging to q-bio in our dataset. As with OSF, not all records correspond to preprints, as arXiv allows uploading of both published papers and preprints, with no metadata field to distinguish between the two. And as other preprint servers, arXiv allows users to uploaded new versions of their records. In the database we analysed, more than two thirds of records (n=19,109, 68%) had one version, one fifth of records had two (n=6,187, 22%), and 10% of records had three or more versions (n=2,808).
However, after exploring the metadata in more detail, we learned that not every record in our dataset represents a unique preprint or published paper on arXiv.
Using exact title and author name matching, we identified two sets of duplicate records (0.01%). One of these pairs was a true duplicate (i.e., records one and two), which also contained an admin note for text overlap (more on this below). The other pair (i.e., records three and four) did not contain such a note. In this case, the preprint and its published version had been posted as separate records rather than as different versions of the same record.
Admin notes such as the one described above are added by arXiv moderators to a record’s comments section. As with the example above, they can contain statements about textual overlaps that have been detected between records. Of the 317 (1%) arXiv records in our dataset with admin notes, all but one mentioned a (substantial) text overlap with other records (e.g., example one). The outlier note indicated an: “inappropriate text reuse from external sources” (see Figure 2, below).

According to arXiv documentation, admin notes are not intended to suggest misconduct on the part of the author or that an article does not contain original work. But, as is the case of the example above, they are sometimes used to identify duplicate records. In other cases, we found they were used even in cases of two different publications that build upon previous work of the authors (e.g., example two – records one and two).
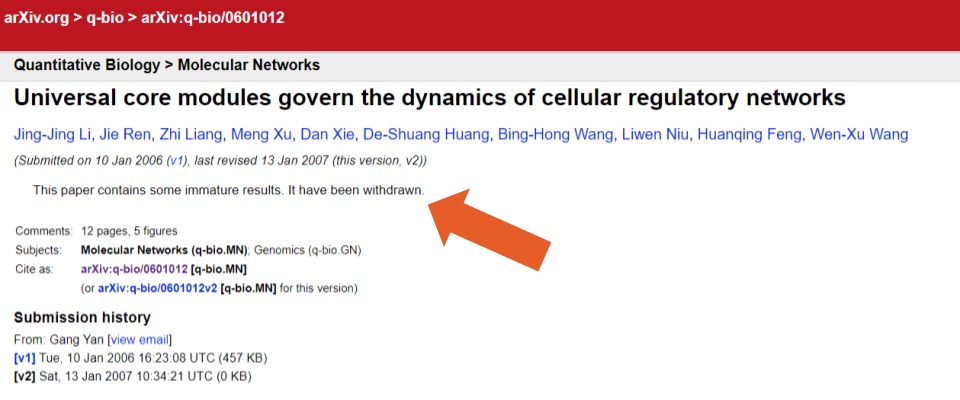
Additionally, as with bioRxiv and the OSF servers, there were no metadata fields for identifying whether a record had been withdrawn and who had posted the withdrawal notice. Instead, we used the comments section (available in the metadata and through this search query) to identify 89 records that used the term “withdrawn” (e.g., examples one and two), and two records that used the term “removed” (e.g., examples three and four). In some cases, however, we also found information about a withdrawal in the abstract text field (e.g. example five, Figure 3 below). In two cases, withdrawn records also contained an admin note (example six, and seven).
Finally, by manually reviewing the comment fields and the abstract fields, we found two additional examples of records that were not unique. In the first case, the comments section of one record stated: “This paper, together with paper q-bio/0607043 are replaced by q-bio/0607043”). In the second, the abstract field had been used to indicate that the record was a supplement to another record.
2) Subjects
As mentioned above, uploaders on arXiv must first specify a section (subject area classification) for their submission, and can then later “cross-list” it to additional sections. The arXiv documentation states that: “It is rarely appropriate to add more than one or two cross-lists”, and that “bad cross-lists will be removed”.)
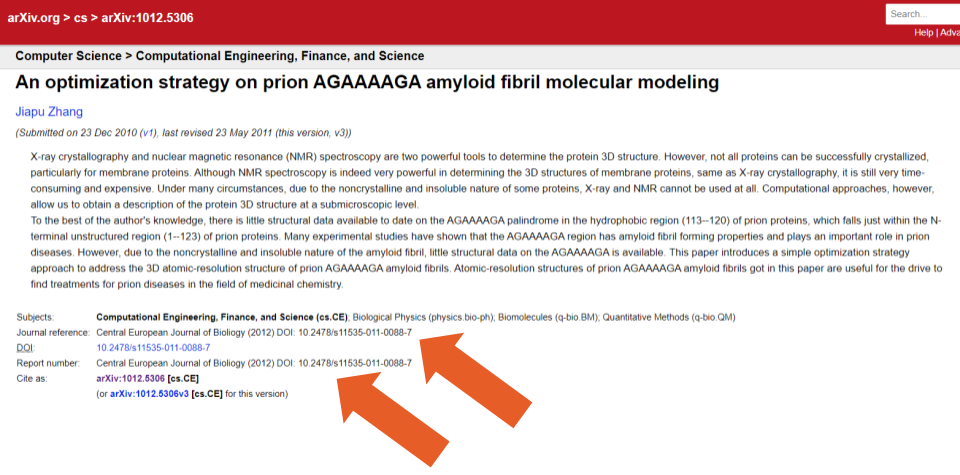
We found that over a quarter of records (n=7,907, 28%) were designated to only one section (not cross-listed). Just over a third of records were cross-listed to one additional arXiv section (n=10,346, 37%), almost a quarter to two (n=6,720, 24%), and eleven percent (n=3,131) to three or more. Of the records that were listed or cross-listed in Quantitative Biology, the most common cross-list was Biological Physics (physics.bio-ph; n=5,113, 18%, see example of a cross-listed record in Figure 4, below).

3) Dates
The arXivRaw metadata format contains two date fields:
- A datestamp, which indicates when the bulk metadata was last updated.
- A submission date, which indicated the date a record was submitted to the server, and is available for each version of a record.
Of the 8,995 records with multiple versions available, 39 had different versions posted on the same day (e.g. version one and two; in this case, a revision was made to adjust the formatting of references). At the other extreme, 235 (0.8%) records included/linked to a revised version posted more than two years later (e.g. version one and two, with 15 years between versions; the only difference between the two versions is that one contains a DOI in the header of the document).
4) Contributors
When uploading records, users enter authors names into a free text field, with a recommendation to separate authors with a comma or an “and”. Because of this, the arXivRaw metadata contains only one contributor field, in which all authors are listed. Affiliation information is neither a part of the submission process, nor does it exist as a separate metadata field. We found, however, that some authors used the free text format of the author field to also include their affiliations (example one and two, Figure 5, below).
Since 2005, arXiv has implemented authority records as means for (co-)authors to confirm or claim authorship and add additional information (e.g., affiliation information, ORCID iDs). In the long term, this should help to identify and map all unique authors of arXiv records. The API for authority records is, however, still in development, and the information it contains is not currently part of the arXivRaw metadata format.
When a consortium is listed as one of the authors, users add the consortium name using the same author free-text field. While this was not the case with the arXivRaw format we used, we noticed that some of the other formats split consortium names into first and last name fields. This introduced errors, as the last word in a consortium’s name was used as a surname and others as forenames (e.g., example one: the arXiv metadata format lists “Consortium” as a last name and “The International Schizophrenia” as forenames).
Finally, arXivRaw metadata format also contains a single text field capturing information about who was the submitter of a record. Using simple exact matching, we identified 12,768 unique submitters, with 4,775 (37%) uploading more than one preprint, and 338 (3%) uploading more than ten. In some of these cases, the name of the uploader differed in spelling or title designation from that used in the record’s author information (e.g., example one, Figure 6, below).

5) Linking preprints to postprints
As with the other preprint servers we studied, arXiv users can link any submission to its published version. They can do so either by inserting a text reference (in the journal reference field), or by adding a DOI (in a DOI field) for the final publication.
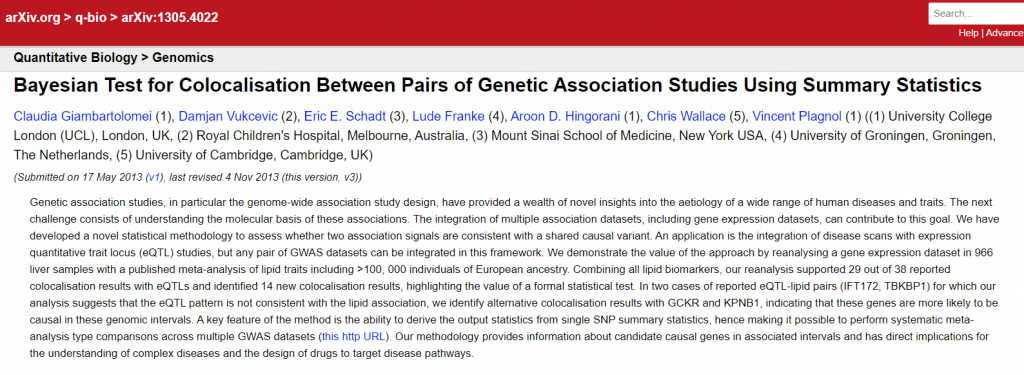
Of the records in the database, 1,517 (5%) included information in a journal reference field, 4,430 (16%) in the DOI field, and 7,991 (28%) in both. Half of the records did not include links to published versions.
However, although links to published versions could suggest that half of the records in the database were preprints that had later been published, it’s difficult to know for certain because arXiv allows users to submit published papers as well, which could also include journal reference or DOI information. With no metadata field or search filter to distinguish preprints from published papers, the numbers above should not be used as representative of the number of preprints on arXiv. Instead, to get such an estimate, researchers should query Crossref or other metadata providers with the DOI and journal reference information for each record, and compare the date of the published records with arXiv submission dates. Of course, this method has its caveats as some preprints are only uploaded after the paper has been published.
Additionally, the journal reference and DOI metadata are not without errors. Despite documentation specifically asking users to provide only a bibliographic reference (without links) in the journal reference field, and despite there being a separate field for DOIs, some users nevertheless entered links (e.g., example one) and DOIs (e.g., example two, Figure 7, below) in the journal reference field.
We found a handful of records that contained duplicate data. A total of 30 (0.1%) records contained a journal reference that corresponded exactly to that of another record, and 38 (0.1%) that listed an identical DOI (12 records had both a matching DOI and matching journal reference). Of those we found one case where two different preprints had been combined into a single publication (i.e. records one and two), as well as several instances where the version that had been submitted to a journal was posted as a separate record from the preprint version (e.g., records three and four).
Preprint metadata is dependent on users
So where does this leave us? We found the challenges of working with metadata from arXiv’s q-bio section to be similar to those we encountered within OSF and biorXiv. However, most of the errors in the data seemed to have stemmed from the way the metadata entry fields were used during preprint upload. To reduce errors like this in future, we recommend that preprints servers support their users by checking and validating the manual inputs used to generate metadata information.
References
- Ginsparg P. ArXiv at 20. Nature. 2011;476(7359):145.
- Feldman S, Lo K, Ammar W. Citation Count Analysis for Papers with Preprints. arXiv preprint arXiv:180505238. 2018.
- Sutton C, Gong L. Popularity of arXiv.org within Computer Science. arXiv preprint arXiv:171005225. 2017.
- Wang Z, Glänzel W, Chen Y, editors. How self-archiving influences the citation impact of a paper. Proceedings of the 23rd International Conference on Science and Technology Indicators; 2018: CWTS, Leiden University; Leiden.
- Larivière V, Sugimoto CR, Macaluso B, Milojević S, Cronin B, Thelwall M. arXiv E‐prints and the journal of record: An analysis of roles and relationships. Journal of the Association for Information Science and Technology. 2014;65(6):1157-69.
Note: We shared a draft of our blog post with the arXiv team prior to its publication and incorporated their feedback. Comments? Questions? Drop us a line on Twitter (tag #scholcommlab).
1 Comment
Comments are closed.
[…] In this post, we draw from our experiences working with metadata from SHARE, OSF, bioRxiv, and arXiv to provide four basic recommendations for those building or managing preprint […]