ScholCommLab co-director receives funding to analyze patterns of how research data is cited and reused
Stefanie Haustein wins Alfred P. Sloan Foundation research grant of $200K US
Have you ever wondered what motivates researchers to reuse open data and what makes them cite (or not cite) datasets in their work? Or how sharing, reusing and citing open data differs between research areas or changes during a researcher’s career?
ScholCommLab co-director Stefanie was awarded $199,929 US ($281,660 CDN) by the Alfred P. Sloan Foundation to fund Meaningful Data Counts, an interdisciplinary project exploring a range of questions about scholarly data use and citation. Together with co-PI Isabella Peters, Professor of Web Science at the ZBW Leibniz Information Centre for Economics and CAU Kiel University (Germany), Stefanie and her team will conduct the basic research necessary to understand how datasets are viewed, used, cited, and remixed.
“Many researchers have to publish a paper to receive credit for collecting, cleaning, and curating scientific data, because datasets don’t count when applying for a tenure-track job or promotion.”
“Many researchers have to publish a paper to receive credit for collecting, cleaning, and curating scientific data, because datasets don’t count when applying for a tenure-track job or promotion,” Stefanie explains. While an increasing number of journals and funding agencies now require data to be open, research datasets are still considered second-class research contributions. By documenting the uptake and reuse of data across disciplines, she hopes the project will help overcome this perception and incentivize open research practices. “Data metrics that capture dataset views, downloads and citations might help to showcase the value of open data and contribute to elevate its status to first-class scholarly output,” she says.
On the other hand, Stefanie acknowledges that metrics could harm the scientific system in the long run, replicating some of the adverse effects associated with current bibliometric indicators. Therefore, a primary goal of the project is to support the creation of meaningful multidimensional metrics instead of promoting raw numbers and crude rankings. “We want to support the evidence-based development of metrics that are appropriate and well thought-out instead of repeating mistakes of the past by creating more flawed indicators, like the h-index and impact factor,” she explains.
The project team—which, in addition to Stefanie and Isabella, includes collaborators Daniella Lowenberg, Felicity Tayler, Rodrigo Costas, Peter Kraker, Philippe Mongeon, and Nicolas Robinson-Garcia—will therefore take a mixed-methods approach to explore how datasets are (re)used and cited across disciplines. Using metadata provided by DataCite, it will build on ScholCommLab member Asura Enkhbayar’s doctoral research to develop a theory of data citations. As part of the larger Make Data Count (MDC) team, the researchers will work in close collaboration with MDC's infrastructure and outreach specialists. Also funded by Sloan, this project aims to build a bibliometrics dashboard and drive adoption of standardized practices.
“We are thrilled to expand our Make Data Count team and scope to include the bibliometrics community,” says Daniella Lowenberg, the project’s PI and Data Publishing Product Manager at the California Digital Library. Martin Fenner, Technical Director at DataCite and MDC technical lead, agrees: “Going forward, Stefanie and her colleagues will provide us with important insights into researcher behavior towards open data,” he says. “I am excited to see Make Data Count transition into the next phase.”
During the two-year funding period the grant will support two postdoctoral researchers—one at the University of Ottawa (quantitative) and one at Isabella’s Web Science research group at the ZBW. “I am very happy to join forces with the ScholCommLab and strengthen the German-Canadian research connection,” she says. “After having worked together with Stefanie for more than a decade—we studied together in Düsseldorf—this is our first shared research grant, which is really exciting.”
Stefanie agrees: "There's a lot of research to be done, but I'm hopeful that, together with MDC, this project will make a real difference in how the scholarly community values open data."
Interested in learning more? For updates and job postings about the Meaningful Data Counts project, sign up for the ScholCommLab’s newsletter.
Analyzing preprints: The challenges of working with metadata from arXiv’s Quantitative Biology section
By Mario Malički, Maria Janina Sarol, and Juan Pablo Alperin
This blog post is the last of a four part series documenting the methodological challenges we faced during our project investigating preprint growth and uptake. Stay tuned for our next post, in which we’ll share general recommendations and hopes for the future of preprint metadata.
Following our analyses of SHARE, OSF, and bioRxiv metadata, (which you can read about here, here, and here), we went on to explore the metadata of arXiv’s Quantitative Biology section.
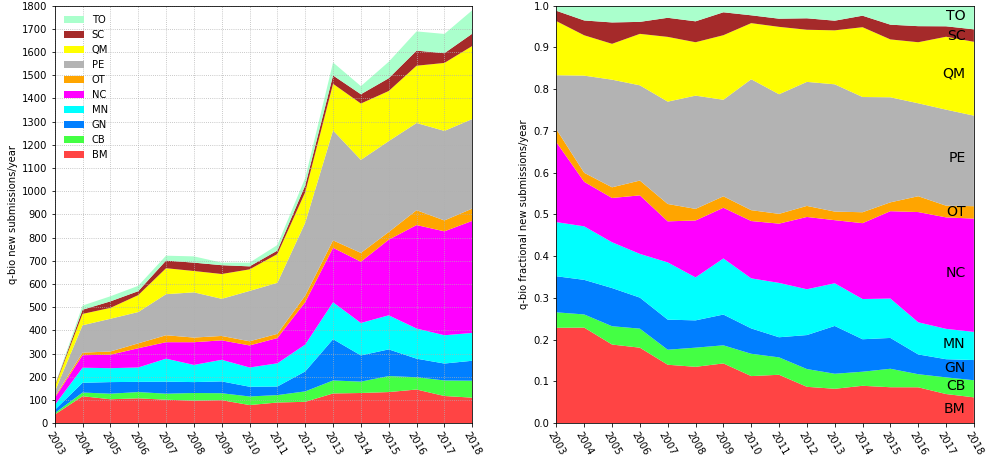
Started in 1991 as a server for theoretical high-energy physics preprints, arXiv has since expanded to cover many other disciplines and has become the largest repository of preprints in science.1 Its growth has made it an important part of research communication in many fields. Research has shown that usage of arXiv has increased dramatically in the last 10 years in computer science,2-3 as well as in library and information science.4 Research also showed that, across all records and disciplines, 64% of arXiv preprints are eventually published in journals indexed in Web of Science (WoS), and, in turn, journals in WoS are increasingly citing arXiv records.5 arXiv subsection dedicated to Quantitative Biology started in 2003 (Figure 1, below).
How does q-bio work?
ArXiv differs from other servers we’ve analyzed in some key ways. First, unlike submissions uploaded to OSF and bioRxiv, arXiv preprints are not assigned a DOI, but an arXiv identifier. In addition, although all uploaders on arXiv (whether they are authors or third-party submitters) must assign their submission to a primary section (e.g., q-bio), and they can also subsequently “cross-list” it to other arXiv sections to increase their audience.
On September 3, 2019, we downloaded the metadata for all arXiv records that were listed or cross-listed to q-bio using arXiv’s Open Archives Initiative Protocol for Metadata Harvesting compatible API. Unlike the metadata of SHARE, OSF, bioRxiv, and Rxivist, which is distributed only in JSON, arXiv metadata can be obtained in four XML formats. We based our analyses on arXivRaw — the format that most closely matches arXiv’s internal format. We then explored the same metadata fields we investigated for SHARE, OSF, and bioRxiv (see our previous posts). Additional details of the various metadata fields available can be found on arXiv’s help pages.
1) Records
There were 28,104 records categorized as belonging to q-bio in our dataset. As with OSF, not all records correspond to preprints, as arXiv allows uploading of both published papers and preprints, with no metadata field to distinguish between the two. And as other preprint servers, arXiv allows users to uploaded new versions of their records. In the database we analysed, more than two thirds of records (n=19,109, 68%) had one version, one fifth of records had two (n=6,187, 22%), and 10% of records had three or more versions (n=2,808).
However, after exploring the metadata in more detail, we learned that not every record in our dataset represents a unique preprint or published paper on arXiv.
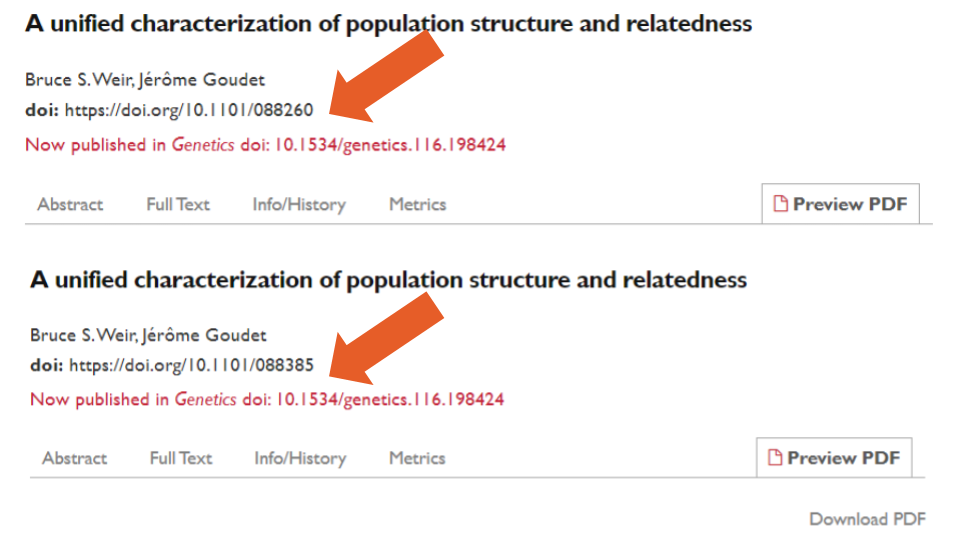
Using exact title and author name matching, we identified two sets of duplicate records (0.01%). One of these pairs was a true duplicate (i.e., records one and two), which also contained an admin note for text overlap (more on this below). The other pair (i.e., records three and four) did not contain such a note. In this case, the preprint and its published version had been posted as separate records rather than as different versions of the same record.
Admin notes such as the one described above are added by arXiv moderators to a record’s comments section. As with the example above, they can contain statements about textual overlaps that have been detected between records. Of the 317 (1%) arXiv records in our dataset with admin notes, all but one mentioned a (substantial) text overlap with other records (e.g., example one). The outlier note indicated an: “inappropriate text reuse from external sources” (see Figure 2, below).

According to arXiv documentation, admin notes are not intended to suggest misconduct on the part of the author or that an article does not contain original work. But, as is the case of the example above, they are sometimes used to identify duplicate records. In other cases, we found they were used even in cases of two different publications that build upon previous work of the authors (e.g., example two – records one and two).
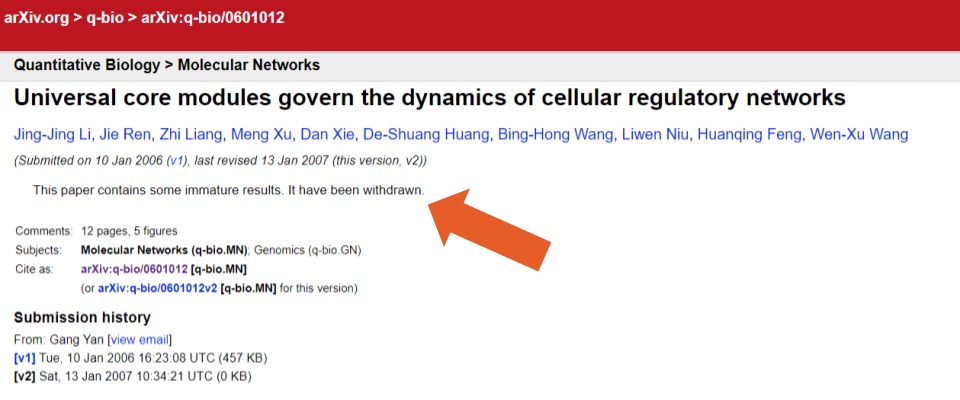
Additionally, as with bioRxiv and the OSF servers, there were no metadata fields for identifying whether a record had been withdrawn and who had posted the withdrawal notice. Instead, we used the comments section (available in the metadata and through this search query) to identify 89 records that used the term “withdrawn” (e.g., examples one and two), and two records that used the term “removed” (e.g., examples three and four). In some cases, however, we also found information about a withdrawal in the abstract text field (e.g. example five, Figure 3 below). In two cases, withdrawn records also contained an admin note (example six, and seven).
Finally, by manually reviewing the comment fields and the abstract fields, we found two additional examples of records that were not unique. In the first case, the comments section of one record stated: “This paper, together with paper q-bio/0607043 are replaced by q-bio/0607043”). In the second, the abstract field had been used to indicate that the record was a supplement to another record.
2) Subjects
As mentioned above, uploaders on arXiv must first specify a section (subject area classification) for their submission, and can then later “cross-list” it to additional sections. The arXiv documentation states that: “It is rarely appropriate to add more than one or two cross-lists”, and that “bad cross-lists will be removed”.)
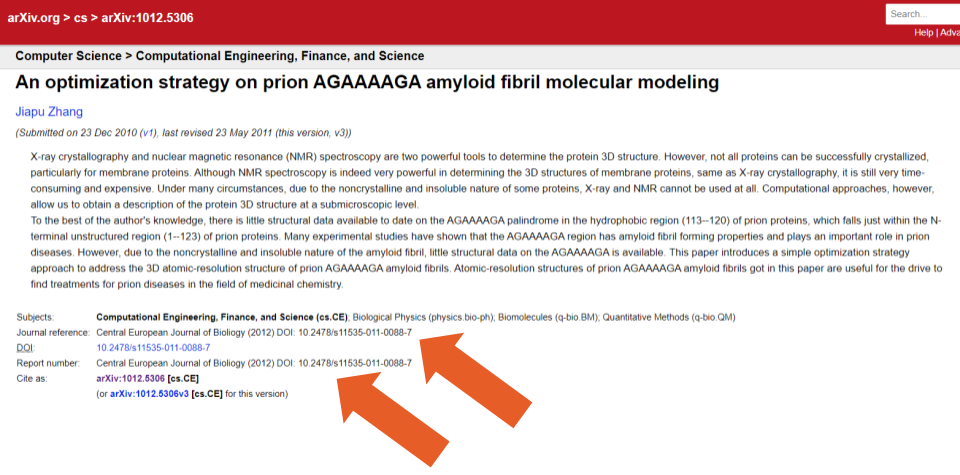
We found that over a quarter of records (n=7,907, 28%) were designated to only one section (not cross-listed). Just over a third of records were cross-listed to one additional arXiv section (n=10,346, 37%), almost a quarter to two (n=6,720, 24%), and eleven percent (n=3,131) to three or more. Of the records that were listed or cross-listed in Quantitative Biology, the most common cross-list was Biological Physics (physics.bio-ph; n=5,113, 18%, see example of a cross-listed record in Figure 4, below).

3) Dates
The arXivRaw metadata format contains two date fields:
- A datestamp, which indicates when the bulk metadata was last updated.
- A submission date, which indicated the date a record was submitted to the server, and is available for each version of a record.
Of the 8,995 records with multiple versions available, 39 had different versions posted on the same day (e.g. version one and two; in this case, a revision was made to adjust the formatting of references). At the other extreme, 235 (0.8%) records included/linked to a revised version posted more than two years later (e.g. version one and two, with 15 years between versions; the only difference between the two versions is that one contains a DOI in the header of the document).
4) Contributors
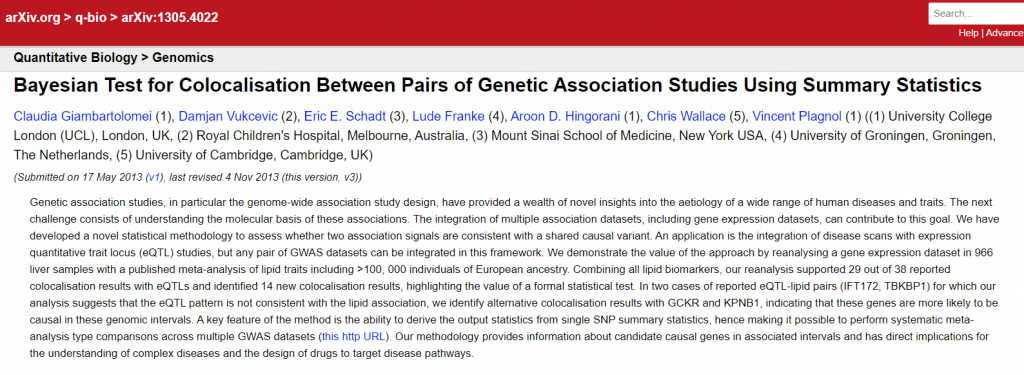
When uploading records, users enter authors names into a free text field, with a recommendation to separate authors with a comma or an “and”. Because of this, the arXivRaw metadata contains only one contributor field, in which all authors are listed. Affiliation information is neither a part of the submission process, nor does it exist as a separate metadata field. We found, however, that some authors used the free text format of the author field to also include their affiliations (example one and two, Figure 5, below).
Since 2005, arXiv has implemented authority records as means for (co-)authors to confirm or claim authorship and add additional information (e.g., affiliation information, ORCID iDs). In the long term, this should help to identify and map all unique authors of arXiv records. The API for authority records is, however, still in development, and the information it contains is not currently part of the arXivRaw metadata format.
When a consortium is listed as one of the authors, users add the consortium name using the same author free-text field. While this was not the case with the arXivRaw format we used, we noticed that some of the other formats split consortium names into first and last name fields. This introduced errors, as the last word in a consortium’s name was used as a surname and others as forenames (e.g., example one: the arXiv metadata format lists “Consortium” as a last name and “The International Schizophrenia” as forenames).
Finally, arXivRaw metadata format also contains a single text field capturing information about who was the submitter of a record. Using simple exact matching, we identified 12,768 unique submitters, with 4,775 (37%) uploading more than one preprint, and 338 (3%) uploading more than ten. In some of these cases, the name of the uploader differed in spelling or title designation from that used in the record’s author information (e.g., example one, Figure 6, below).

5) Linking preprints to postprints
As with the other preprint servers we studied, arXiv users can link any submission to its published version. They can do so either by inserting a text reference (in the journal reference field), or by adding a DOI (in a DOI field) for the final publication.
Of the records in the database, 1,517 (5%) included information in a journal reference field, 4,430 (16%) in the DOI field, and 7,991 (28%) in both. Half of the records did not include links to published versions.
However, although links to published versions could suggest that half of the records in the database were preprints that had later been published, it’s difficult to know for certain because arXiv allows users to submit published papers as well, which could also include journal reference or DOI information. With no metadata field or search filter to distinguish preprints from published papers, the numbers above should not be used as representative of the number of preprints on arXiv. Instead, to get such an estimate, researchers should query Crossref or other metadata providers with the DOI and journal reference information for each record, and compare the date of the published records with arXiv submission dates. Of course, this method has its caveats as some preprints are only uploaded after the paper has been published.
Additionally, the journal reference and DOI metadata are not without errors. Despite documentation specifically asking users to provide only a bibliographic reference (without links) in the journal reference field, and despite there being a separate field for DOIs, some users nevertheless entered links (e.g., example one) and DOIs (e.g., example two, Figure 7, below) in the journal reference field.
We found a handful of records that contained duplicate data. A total of 30 (0.1%) records contained a journal reference that corresponded exactly to that of another record, and 38 (0.1%) that listed an identical DOI (12 records had both a matching DOI and matching journal reference). Of those we found one case where two different preprints had been combined into a single publication (i.e. records one and two), as well as several instances where the version that had been submitted to a journal was posted as a separate record from the preprint version (e.g., records three and four).
Preprint metadata is dependent on users
So where does this leave us? We found the challenges of working with metadata from arXiv’s q-bio section to be similar to those we encountered within OSF and biorXiv. However, most of the errors in the data seemed to have stemmed from the way the metadata entry fields were used during preprint upload. To reduce errors like this in future, we recommend that preprints servers support their users by checking and validating the manual inputs used to generate metadata information.
References
- Ginsparg P. ArXiv at 20. Nature. 2011;476(7359):145.
- Feldman S, Lo K, Ammar W. Citation Count Analysis for Papers with Preprints. arXiv preprint arXiv:180505238. 2018.
- Sutton C, Gong L. Popularity of arXiv.org within Computer Science. arXiv preprint arXiv:171005225. 2017.
- Wang Z, Glänzel W, Chen Y, editors. How self-archiving influences the citation impact of a paper. Proceedings of the 23rd International Conference on Science and Technology Indicators; 2018: CWTS, Leiden University; Leiden.
- Larivière V, Sugimoto CR, Macaluso B, Milojević S, Cronin B, Thelwall M. arXiv E‐prints and the journal of record: An analysis of roles and relationships. Journal of the Association for Information Science and Technology. 2014;65(6):1157-69.
Note: We shared a draft of our blog post with the arXiv team prior to its publication and incorporated their feedback. Comments? Questions? Drop us a line on Twitter (tag #scholcommlab).
OAEvolution: Trends in Open Access and Scholarly Communications
 This week marks the 10 year anniversary of Open Access Week—a global event that strives "to make Open Access a new norm in scholarship and research." In celebration of this momentous occasion, the University of Ottawa Library asked ScholCommLab co-director Stefanie Haustein about recent trends in open access and scholarly communication. In this interview with the Library's Communication Advisor Meryem El Marzouki, Stefanie reflects on the past, present, and future of OA in our changing scholarly communication landscape.
This week marks the 10 year anniversary of Open Access Week—a global event that strives "to make Open Access a new norm in scholarship and research." In celebration of this momentous occasion, the University of Ottawa Library asked ScholCommLab co-director Stefanie Haustein about recent trends in open access and scholarly communication. In this interview with the Library's Communication Advisor Meryem El Marzouki, Stefanie reflects on the past, present, and future of OA in our changing scholarly communication landscape.
uOttawa Library: How do you see the evolution of open access over the last 5 years?
SH: In the last year, Plan S has certainly shaken up academic publishing. A group of Research Funding and Research Performing Organizations in Europe introduced an ambitious roadmap to make all funded research outputs OA. The fact that Plan S does not allow hybrid OA has been seen by many as a (too) radical change. Since publishers make the scholarly community pay twice when they charge hybrid APCs for OA publication in subscription journals, I think it is a very convincing and very much needed regulation, considering that large publishing houses like Elsevier continue to make profits higher than Apple and Big Pharma on the shoulders of academia.
The extent to which scholarly articles are published OA has continually increased over the last years. A major study conducted for the European Union reported that since 2007 more than 50% of peer-reviewed journal articles are available OA. More conservative estimates report that 45% of articles published in 2015 are OA.
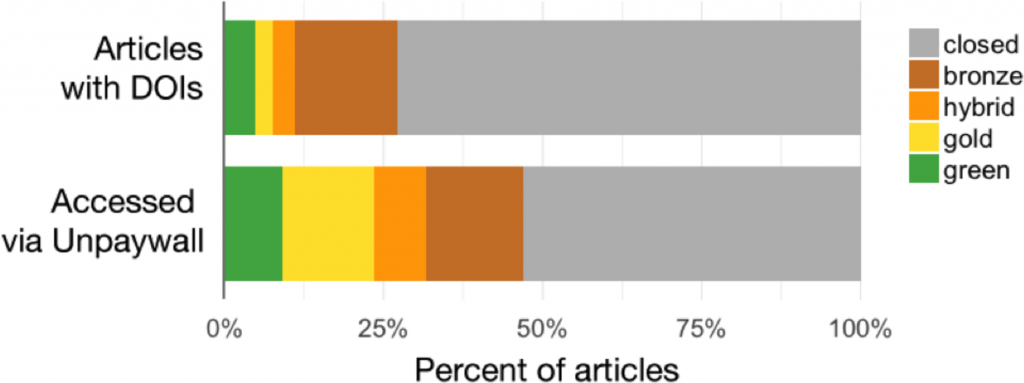
However, what is considered OA beyond green and gold depends on who you ask: Should copies of papers on academic social networks, which often infringe copyright, be considered OA? Should documents on illegal platforms such as SciHub, so-called "black OA," be included? Or are only articles with a specific free to read and free to reuse license truly OA? A recent study added another colour to the spectrum: bronze OA, for articles that are “free to read on the publisher page, but without any clearly identifiable license”.
uOttawa Library: What is new/special to highlight about open access?
SH: When, in October 2015, the editorial board of the Elsevier journal Lingua resigned over the inability to agree to switch to an OA model and subsequently founded the new OA journal Glossa, a precedent was set for other editorial boards to consider changing to OA. Since then the boards of at least two journals followed suit and left their publisher to flip to OA. I am proud to be part of the former editorial board of the Journal of Informetrics (Elsevier), which stepped down unanimously in January 2019 to found the new OA journal Quantitative Science Studies (MIT Press).
But not all publishers seem resistant to change. One of the latest trends are so-called "transformative agreements," where publishers agree to make journals OA by shifting from a reader-pays (library subscriptions) to an author-pays model (gold OA with APCs) in an agreed-upon time frame. Much of these agreements to flip journals are also based on Plan S requirements, which stipulate that subscription journals need to fully transition to OA by 2023 in order to be compliant.
uOttawa Library: Do you have any idea of what the future might hold for open access?
SH: I would hope that the scholarly community takes back ownership and control over scholarly publishing by flipping journals and demanding OA publication with affordable and transparent APCs. Taking back control of scholarly communication infrastructure does not stop at OA publishing but extends to open data and open citations.
"I would hope that the scholarly community takes back ownership and control over scholarly publishing by flipping journals and demanding OA publication with affordable and transparent APCs."
Scholars need to understand that without the scholarly community submitting, reviewing, and editing manuscripts, a journal is nothing but a fancy website!
This interview was conducted and edited by Meryem El Marzouki, Communication Advisor at the University of Ottawa Library. If you are in Ottawa, check out and participate in uOttawa Library’s OA week events.
Analyzing Preprints: The challenges of working with metadata from bioRxiv
By Mario Malički, Maria Janina Sarol, and Juan Pablo Alperin
This blog post is the third in a four part series documenting the methodological challenges we faced during our project investigating preprint growth and uptake. Stay tuned for our final post, in which we’ll dive into the arXiv metadata.

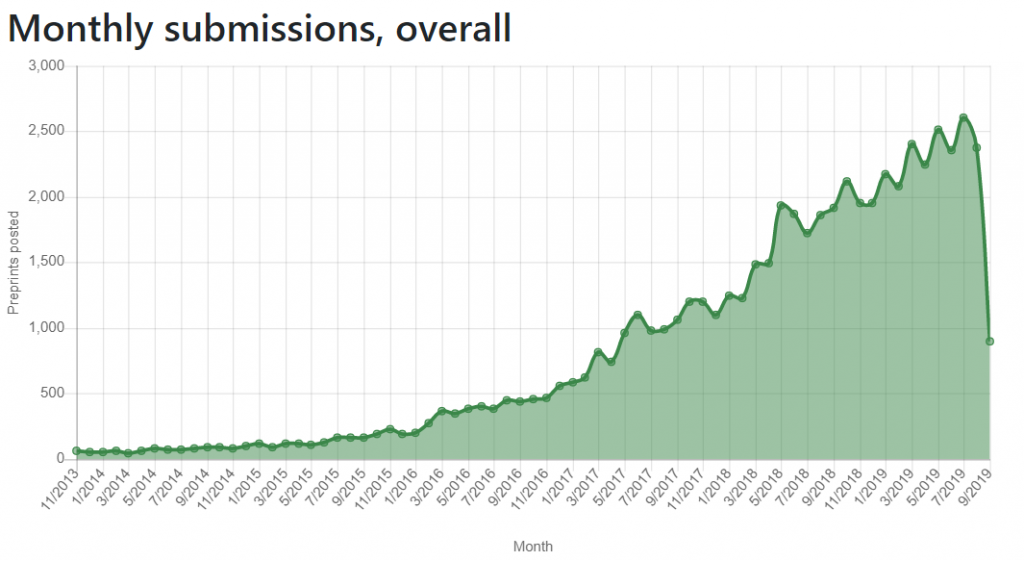
Following our analysis of the challenges working with SHARE and OSF metadata (read about them in our previous blogs here and here), we explored the metadata of bioRxiv—a preprint server focused on biology research. Since its inception in 2013, bioRxiv has received significant attention,1 and became the largest biology-focused preprint server (Figure 1, below). Studies showed that posting a preprint on bioRxiv was associated with more citations and higher Altmetric scores than articles that were not posted as preprints.2-4 Additionally, recent research (itself posted on bioRxiv) has shown slight differences between preprints posted on bioRxiv and postprints published in journals indexed in PubMed, with peer reviewed articles having better reporting of reagents (i.e., drug suppliers and antibody validation) and experimental animals (i.e., reporting of strain, sex, supplier and randomization), but bioRxiv articles having better reporting of unit-level data, completeness of statistical results, and exact p-values.5
Unlike OSF servers we previously analysed, bioRxiv only accepts unpublished/unrefereed manuscripts that have not been peer reviewed or accepted for publication in a journal (Figure 1, below). Furthermore, bioRxiv specifically states it does not accept theses, dissertations, or protocols. It accepts only preprints written in English and uploaded as either Word documents or PDFs. Moreover, all uploaded preprints, including their updated versions, undergo a basic screening process for content that is offensive, non-scientific, or that might pose a health or biosecurity risk, and undergo iThenthicate plagiarism check and duplicate search (information provided by bioRxiv). To encourage use of preprints, bioRxiv allows users to directly transmit uploaded documents and their metadata to a number of journals, and journals to deposit received manuscripts as preprints to bioRxiv.
Despite not yet releasing their own public API, bioRxiv metadata can be retrieved from Crossref and through the Rxivist site. BioRxiv deposits metadata to Crossref as part of the official scholarly record; while the metadata available from Rxivist is scraped from the bioRxiv website.6
For our analysis, we downloaded metadata from June 18 to 20, 2019 for all posted content in Crossref—that is, all “preprints, eprints, working papers, reports, dissertations, and many other types of content that has been posted but not formally published”—and filtered it for records from bioRxiv. Guided by a description of Crossref metadata fields, we then set out to explore the same basic metadata fields as we did for SHARE and OSF. Here’s what we found:
1) Records
Our final database contained 52,531 preprints. Although bioRxiv users have to categorize their preprints into one of three categories (New Results, Confirmatory Results, or Contradictory Results), this information is only available through the biorXiv search engine—and not in the Crossref or Rxivist metadata. On August 28, 405 (0.7%) preprints were listed as Contradictory and 1,090 (1.9%) as Confirmatory; all the rest were classified as New Results.
Although records can be removed in cases of discovered plagiarism or breach of relevant ethical standards (see the bioRxiv Submission Guide for details), there was no metadata field for withdrawn status in either Crossref or Rxivist. However, the bioRxiv’s search engine includes a filter option that on August 28 listed 49 preprints as withdrawn. Unlike for published papers, for which the Committee on Publication Ethics recommends keeping the full text of the record available after indicating its retracted or withdrawn status, some uploaded documents of withdrawn preprints in bioRxiv, are removed from the server, and their abstracts replaced with removal reason statements. This is similar to the practice we found in OSF.
There is also no metadata information available for authors of the removal statements, although, in some cases, the statements themselves contain information about the author (e.g., as in example one, a preprint removed by the bioRxiv moderators, or example two, a preprint removed at the request of the authors). However, in several cases the reason for removal is not clear (e.g., example three, Figure 2 below).

Using Crossref Metadata, we also identified 10 duplicate cases, where the title and author(s) of one preprint was an exact match to that of one or more other preprints (e.g., example one, preprints one and two are identical; example two, preprints one, two, and three are different versions of the same preprint). It is worth nothing that bioRxiv has recently implemented a duplicate search.
2) Subjects
On upload, bioRxiv users have to assign their preprint into one of 25 subject areas. Previously users could assign preprints to two additional subject areas – Clinical Trials and Epidemiology – until content in these areas were redirected to medRxiv, a preprint server for clinical sciences launched in June. Taking these two discontinued subject areas into account, we originally expected the bioRxiv metadata to include 27 subject area classifications. Instead, the Crossref metadata listed 29 areas, including two areas not listed on the bioRxiv website: Scientific Communication (n=16) and Pharmacology (n=26), which existed alongside listed Scientific Communication and Education (n=322), and Pharmacology and Toxicology (n=400) areas. (A recent update of bioRxiv metadata has corrected this.)
There were only eight records (0.02%) which did not have a subject information in the Crossref metadata. Of these eight, only two listed subject information on the bioRxiv website and in Rxivist (i.e., records one and two); the remaining six did not (i.e., records one, two, three, four, five, and six).
3) Dates
There are six date fields in the Crossref metadata for bioRxiv preprints: indexed, posted, issued, accepted, created, and deposited. All six of these fields were populated for each of the bioRxiv preprints in our database.
Crossref defines these fields as follows:
- Date indexed: when the metadata was most recently processed by Crossref;
- Date posted: when the content was made available online;
- Date issued: the earliest known publication date. This was equal to date posted in all the cases we studied;
- Date accepted: the date on which a work was accepted for publication. For 37,730 (72%) records, this was equal to the date posted/issued, for 13,517 (26%), it preceded the date posted, and for 1290 (2%) it succeeded it. A description of why this is so, would be a welcome addition to the metadata documentation;
- Date created: when the DOI was first registered. For all but one preprint, Figure 3 below, this date was the same or succeeded the date posted/issued—likely because most DOIs are assigned on the same day or a few days after a preprint is created on the bioRxiv server.
- Date deposited: when the metadata was last updated (by bioRxiv).
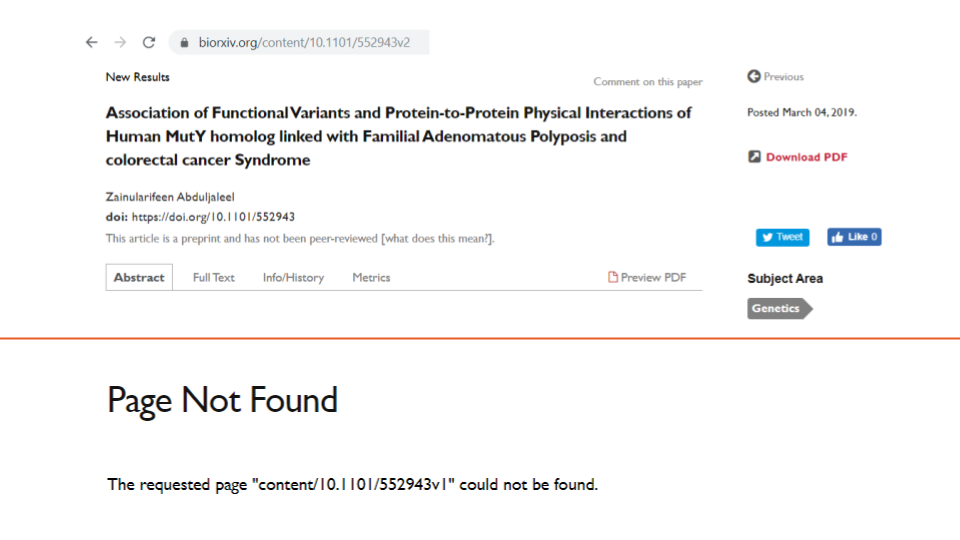
The Rxivist metadata contains only one date field, the date when a preprint was first posted. Metadata regarding version numbers or update dates is not available from either Crossref or Rxivist. However, it is possible to discern the version number by looking at the last two characters of the biorxiv_url field in Rxivist (e.g., example one, whose biorxiv_url ends with v7).
4) Contributors
When entering author information during preprint upload at bioRxiv, users are required to fill in each author’s given and family name and institutional affiliation. They can also optionally fill in the author’s middle name, middle name initials, name suffix, home page, and ORCID iD. Alternatively, users can choose to add a Collaborative Group/Consortium as an author, by filling in a free text field. This information, alongside the uploaded document, is then used to create a final pdf version of the preprints (which users have to approve), and from which the final metadata is extracted.
Only three records in the Crossref metadata lacked any author information (specifically, records one, two, and three—this third one has since been corrected). All other records included, at minimum, at least one consortium or family name. We found 24 (0.05%) records with an empty given name field. In some cases this was because a consortium name had been entered as a family name (e.g., example one), in others because only a family name had been entered (e.g., examples two and three). Additionally, we identified 33 records where an author’s full name (given and family name together) exactly matched that of another author on the same record (these errors have since been corrected). For 859 (2%) preprints, we found a Collaborative Group/Consortium as one of the authors. Two of these (records one and two) were exclusively authored by a consortium. Of preprints with consortia authors, 156 (18%) listed more than one Consortium. One preprint (see Figure 4, below), listed the same consortium name twice.

We also identified cases where authors names were entered as consortia (e.g., example three), as well as cases where the consortia field was used to enter “zero”, “No”, “None”, or “N/A” (many of these issues have since been corrected). In a handful of cases, different records used different spellings or acronyms for the same consortia (e.g., the Karolinska Schizophrenia Project (KASP) was also spelled as Karolinska Schizophrenia Project KaSP, KaSP, and KaSP Consortium). All of these issues make it difficult to accurately determine the number of unique consortia listed as co-authors.
In the Crossref metadata, each author is assigned a Sequence field identifying them as either the first or an additional author on a preprint. As expected, in almost all cases, the additional authors appeared in the metadata in the same order as on the bioRxiv webpage. However, in Crossref JSON metadata format consortia always occupy the last spot of the author list (e.g. example one, where a consortia is the middle position of the author byline, but last in the metadata), which makes it impossible to use Crossref JSON metadata format to determine byline order for preprints co-authored with consortia. Crossref XML metadata format and Rxivist metadata retains the correct author order (note: they do not contain a field indicating exact byline position).
For 28,663 (55%) preprints in Crossref metadata, at least one author had an ORCID iD; for 4,177 (8%), all of the authors did. For 13 preprints, two or more authors shared the same ORCID iD (e.g., example one and two).
Lastly, although bioRxiv requires uploaders to add affiliation information for all authors during upload, this information isn’t captured in the Crossref metadata (though it is available in Rxivist).
5) Linking preprints to postprints
As Indicated in our previous blogposts, it is important to identify if a record on a preprint server is actually a preprint. BioRxiv monitors title and authors of records in Crossref and PubMed. On identifying a potential match, they send an email to the corresponding author for confirmation, and then add that information to the preprint metadata. Additionally, users can also manually alert bioRxiv of postprint publications.3
In the Crossref metadata, 20,465 (39%) records had a link to a DOI of their published version. Of those, one record contained its own DOI instead of that of the published postprint (at the time of writing this error had been corrected on the bioRxiv site but not in the metadata deposited to Crossref). We found an additional 29 pairs of preprints that shared an identical postprint DOI with each other. In some cases, this was caused by different versions of the same preprint being uploaded to bioRxiv as separate preprints (e.g., example one: preprints one and two, example two: preprints three and four). In others, the same version had been uploaded twice as two separate preprints (e.g., example three: preprints five and six, Figure 5, below). Additionally, despite accepting only original research preprints, both review papers and published papers have been found among bioRxiv records6-7 (discussion of the preprint community and bioRxiv moderators about these issues can be found here).
Finally, five preprints contained links to reviews and endorsements received on F1000 Prime in Crossref’s relation metadata field (e.g., example one).
Metadata continues to improve
In the two months that have passed since we first downloaded the bioRxiv Crossref metadata and writing of this post, the bioRxiv team has been working and improving the metadata they provide. We hope that they similarly address the errors and omissions we’ve identified above, and enrich the metadata with information about author affiliations, withdrawn records, article categorization, versions, and comments. Overall, we identified fewer issues with bioRxiv metadata than we did for SHARE and OSF. We hope researchers using bioRxiv metadata from either Crossref or Rxivist will report if and how they overcame the challenges we described.
References
- Kaiser J. Are preprints the future of biology? A survival guide for scientists. Science. 2017;397.
- Fu DY, Hughey JJ. Releasing a preprint is associated with more attention and citations. bioRxiv. 2019:699652.
- Fraser N, Momeni F, Mayr P, Peters I. The effect of bioRxiv preprints on citations and altmetrics. bioRxiv. 2019:673665.
- Serghiou S, Ioannidis JP. Altmetric Scores, Citations, and Publication of Studies Posted as Preprints. JAMA: The Journal of the American Medical Association. 2018;319(4):402-4.
- Carneiro CFD, Queiroz VGS, Moulin TC, Carvalho CAM, Haas CB, Rayêe D, et al. Comparing quality of reporting between preprints and peer-reviewed articles in the biomedical literature. bioRxiv. 2019:581892.
- Abdill RJ, Blekhman R. Meta-Research: Tracking the popularity and outcomes of all bioRxiv preprints. eLife. 2019;8:e45133.
- Anaya J. 2018. [cited 2019]. Available from: https://medium.com/@OmnesRes/the-biorxiv-wall-of-shame-aa3d9cfc4cd7.
Comments? Questions? Drop us a line on Twitter (tag #scholcommlab). We have shared the issues outlined above with the bioRxiv team, and incorporated their feedback into this post. Additionally, recently bioRxiv has released a beta version of their own API. We welcome your insights and experiences with working with preprint metadata. Our source code is available on GitHub.
Analyzing preprints: The challenges of working with OSF metadata
By Mario Malički, Maria Janina Sarol, and Juan Pablo Alperin
This blog post is the second in a four part series documenting the methodological challenges we faced during our project investigating preprint growth and uptake. Stay tuned for our next post, in which we’ll describe our experiences with the bioRxiv metadata.
Our analysis of the SHARE database, described in our previous blog post, revealed many challenges of working with metadata that was merged from different preprint servers. To continue with our research, we needed to proceed with a detailed analysis of one server (provider) at a time. We decided to start with preprint servers created by, or in partnership with, The Center for Open Science (COS). Using their Open Science Framework (OSF) preprints infrastructure, COS launched its first interdisciplinary preprint server, titled OSF Preprints, in 2016, and in the years following helped launch additional 25 servers (Figure 1 below), each tailored for a specific discipline or country. We downloaded all 29,430 records available through the OSF API on June 18, 2019 and explored the main metadata fields, just as we had done for SHARE (documentation on OSF server metadata fields is available here).

1) Sources and records per source
While we could easily calculate how many records existed on each of the 25 servers, we soon realized that records do not necessarily correspond to preprints. Identifying preprints among the records ended up being more challenging than we had anticipated for a number of reasons.
First, although servers employing the OSF infrastructure can accept different types of records, there is no metadata field (or filter on the OSF search engine) to distinguish between them (SHARE metadata also lacks such a filed). Our exploration revealed that several of the records were postprints (i.e., published, peer reviewed papers (e.g., example one), full conference proceedings containing several papers (e.g., example two), PowerPoint presentations (e.g., example three), or Excel files (e.g., example four). Some of the records were also theses, with one of the servers, Thesis Commons, created specifically for this document type.
Second, we identified a significant number of duplicate records which, if left accounted for, overestimate the number of preprints or records available. By using a strict exact matching of titles and full author names, we found 1,810 (6%) duplicate OSF records. These records corresponded to 778 unique title and author pairs that had been reposted at least once on either the same OSF preprint server (e.g., example one: records one and two, Figure 2, below) or on different servers (example two: records one, two, three, four, and five, or example three: records one and two).

This number, however, underestimates the actual number of duplicates, as users creating these records on OSF servers sometimes enter slightly different information. For example:
- different authors for the same record, as in example four: records one and two;
- different spellings of an author’s name or omission of an author’s middle name, as in example five: records one and two; or
- uploading the same records in two different languages, as in example six: records one and two.
To reliably identify all of the duplicates, we’d need a more detailed comparison of titles, names abstracts, and linked published papers (outlined later in this post).
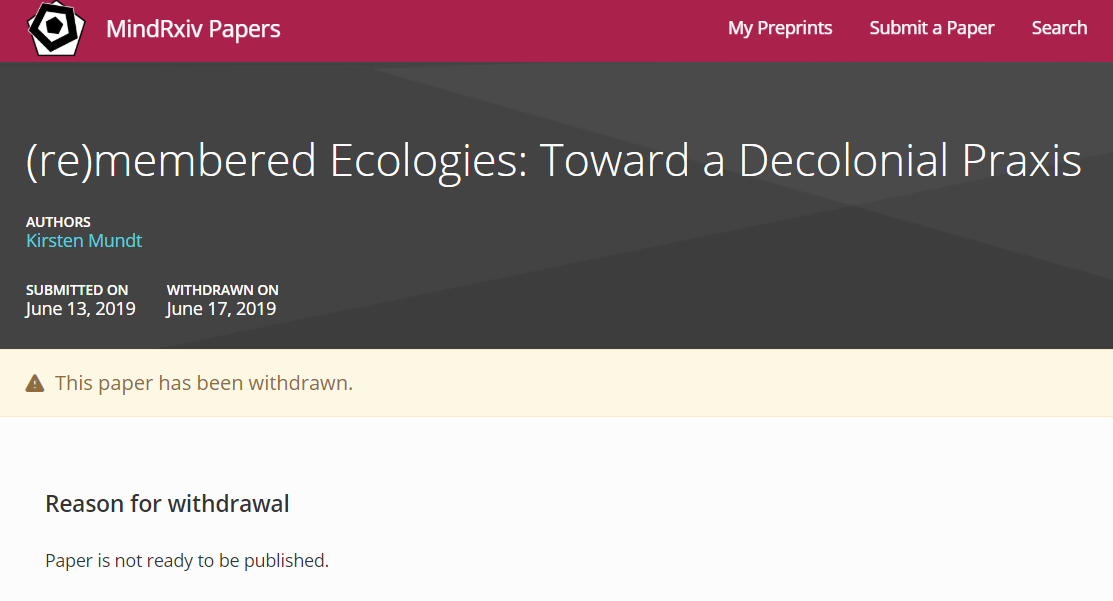
Third, 96 (0.3%) records we identified had been withdrawn at some point. But unlike for published papers, for which the Committee on Publication Ethics recommends keeping the full text of the record available after indicating its retracted or withdrawn status, withdrawn documents on OSF are removed, leaving behind only the webpage and none of the content. This makes it impossible to determine if withdrawn records were indeed preprints or some other kind of document. Reasons for withdrawal were listed on the webpage and available in the metadata for 70 (73%) of records. But while some withdrawal reasons were written by the server moderators (e.g., example one), some by the authors (e.g., example two), there were also cases where it was not clear who wrote them (e.g., example three, Figure 3). No (metadata) information was available on the individuals who provided the reasons for withdrawal.
Additionally, we found six orphaned records, for which the metadata existed in the database, but the webpage was no longer accessible. We could not find explanations on the differences between orphaned and withdrawn records, except for OSF’s orphaned metadata definition, which reads: “A preprint can be orphaned if it's primary file was removed from the preprint node. This field may be deprecated in future versions."
2) Subjects (i.e., scientific discipline or subdiscipline classification)
OSF servers use the bepress discipline taxonomy for classifying records to scientific fields, albeit with slight modifications. For example, bodoArXiv, a server for medieval studies, introduced Medieval Studies as a top level discipline, while the bepress taxonomy has Medieval Studies under the Arts and Humanities top-level discipline. Similarly Meta-Science and Neuroscience were introduced as a top-level disciplines in PsyArXiv, while in bepress Meta-Science does not exist as a separate sub-discipline, and Neuroscience falls under the top-level Life Sciences discipline. Only two of the records we identified did not have subject classification.
3) Dates
OSF metadata contains seven date fields, two of which we planned to use for our analyses: date_published (defined as “the time at which the preprint was published”) and original_publication_date (defined as: “user-entered, the date when the preprint was originally published”). The definition of the other five date fields (date_created, preprint_doi_created, date_withdrawn, date_modified, and date_last_transitioned) is available here. The date_published field appears to be automatically populated, and was therefore present for all records. Original_publication_date, on the other hand, was entered by users for only 9,894 (35%) records.
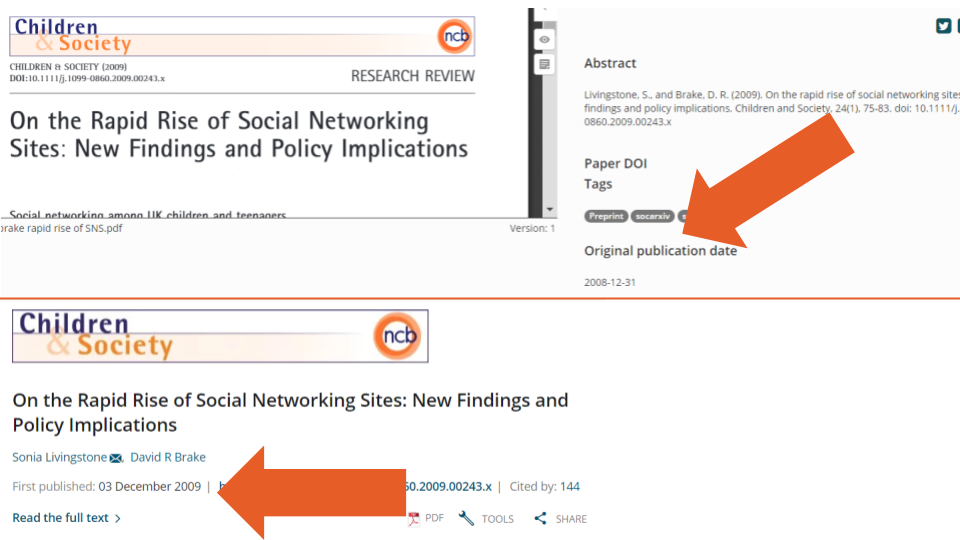
We also found instances where the original_publication_date differed from the date listed in Crossref metadata (e.g., example one, the user entered date is February 13, 2018, but Crossref lists February 14; example two, Figure 4 below, the user entered December 31, 2008, but Crossref lists December 3, 2009). These kinds of discrepancies make it difficult to identify which records correspond to preprints (i.e., by comparing the date when a record was created on a preprint server with the date it was published as a peer reviewed paper). They also pose challenges for determining at what stage of the dissemination process authors uploaded the preprint (i.e., on the same date they submitted a paper to a journal, or at the time of the paper’s acceptance).
4) Contributors
In our previous blog post, we explained how in SHARE, Contributors were divided into Creators (authors) or Contributors (uploaders). In OSF, Contributors are also divided into two similar categories: bibliographic, those that are the authors of the record, and non-bibliographic, those that can upload (new) versions of the records. We found 2,278 (8%) records that had at least one non-bibliographic Contributor (uploader), but only 14 (1%) of those records also listed one of those uploaders as an author. This confirms, like that data in SHARE, that uploaders are rarely also the authors of the records they are uploading, and indicates users are likely misunderstanding the entry fields during preprint uploading.
We identified many of the same issues that plague the Contributor field in SHARE, also in the OSF metadata. When taken together, these issues make the author metadata unreliable for authorship network analysis and other scientific inquiries. For example, we identified cases where the correct number of authors in the metadata differed from the number of authors listed in the uploaded documents (e.g., example one: only one author is listed on the record, but three can be found in the uploaded PDF).
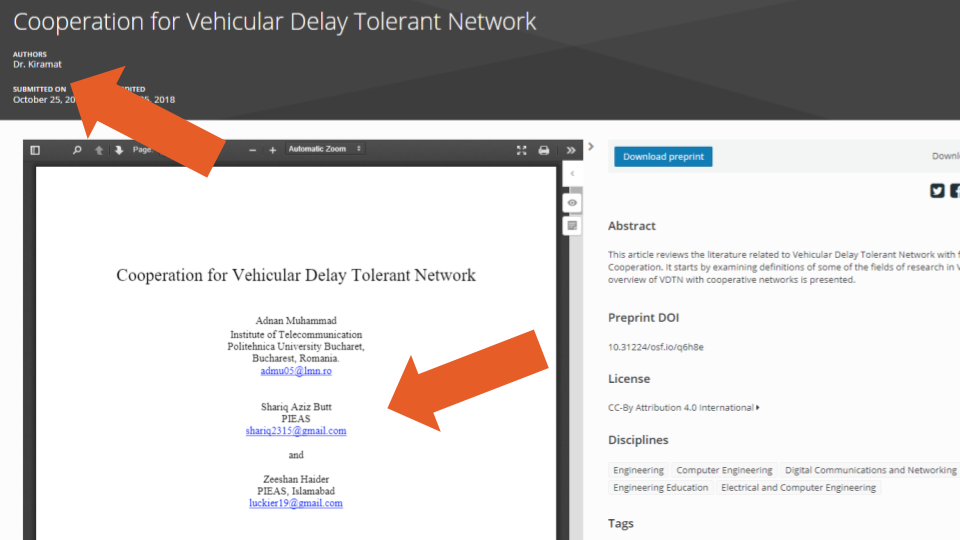
We also encountered cases where Contributors that were specified as non-bibliographic should have been bibliographic (e.g., example two: the metadata lists one bibliographic and five non-bibliographic authors, while the uploaded PDF lists all six as bibliographic authors), as well as cases where the authors listed in the metadata were not among the authors listed in the uploaded PDF (e.g., example three, Figure 5 below: the single author in the metadata differs from the three listed in the uploaded PDF). Additionally, there were 31 (0.1%) records where the same author (using simple exact full name matching) was listed more than once as an author on the same record (e.g., example four).
When we dug deeper into the analyses of authors’ names, we also saw that 137 (0.5%) records were missing an author’s first (given) name or initials (e.g., example one), and 2,091 (7%) were missing a last name (e.g., example two). We also found instances where users listed the name of a journal as the name of the author (e.g., example three, Figure 6 below), the name of a proceeding (e.g., example four) or of an institute (e.g., example five), and cases where the professional titles were included in names (e.g., example six).
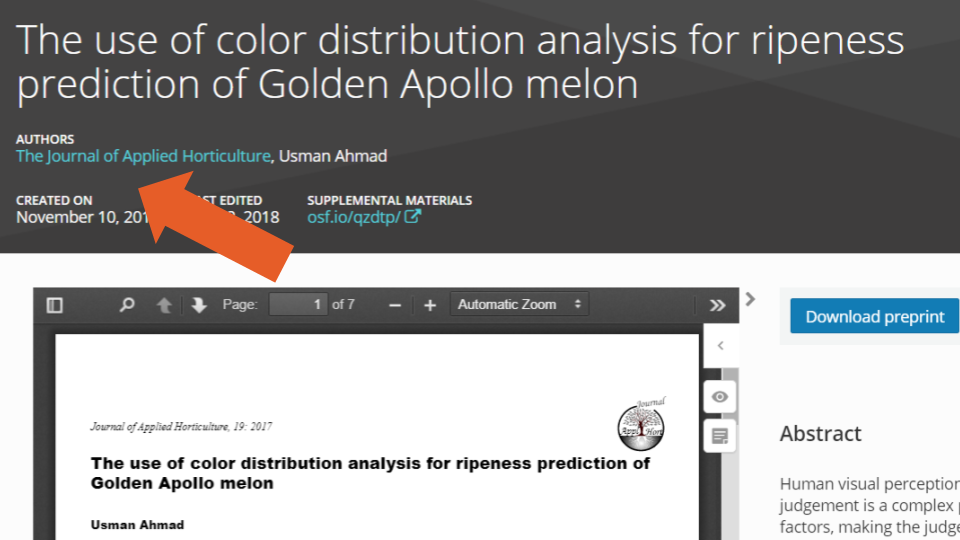
Additionally, like in SHARE, we found inconsistencies regarding the position of authors in the byline order (i.e., their place in the list of authors). We identified 67 cases where two or more authors had the same position in the byline order according to the metadata information, and to the best of our knowledge none of these cases where due to the practice of equal or shared contributorship (e.g., example one: the last two authors share the byline order position according to the metadata, but not according to the uploaded PDF; example two: the first two authors share the byline order position according to the metadata, but not according to the uploaded PDF). Finally, we also identified cases where the byline order in the metadata was completely different than the one in the uploaded document (e.g., example three).
As a final remark on the Contributor field, we noted that information on authors’ affiliations is not (directly) included in the record’s metadata; it is available only for authors who have created an OSF account and have taken the time to fill in their employment information. That is, uploaders who post preprints on the OSF servers are not asked to fill in affiliation information or professional identifiers (e.g., an ORCID iD) for themselves or for their co-authors at the time of the preprint upload. Instead, co-authors are sent an email by the system asking them to create an account, and if they choose to do so, they are then prompted to include their identifiers, education, and professional affiliations (but none of these fields are mandatory). Moreover, because this metadata is linked to the user and not the preprint, it can easily introduce errors and inconsistencies, as affiliations change over time. There were 9,741 (33%) records for which at least one of the co-authors did not create their profile on OSF. Of the 17,510 unique users that did create their profiles, only 3,218 (18%) provided their affiliation information.
Additional exploration of OSF servers
Despite all of the issues with the basic OSF metadata we described above, we still wanted to include OSF servers in our analysis of preprint growth and uptake. To help estimate the number of unique preprints (not records) on the servers, we explored an additional metadata field that we did not explore in SHARE: a record’s link to its published peer reviewed version.
Like many other preprint servers, OSF servers offer a free text field to allow users to link their preprints to their published (likely peer reviewed) version. This can be done at the same time the preprint is uploaded (if the published version already exists), or at any time after it becomes available. However, on OSF, the field is labelled “Peer-reviewed publication DOI”, which means that preprints cannot be linked to papers published in venues that do not assign a DOI. We found 4,965 (17%) records with a peer-reviewed publication DOI.
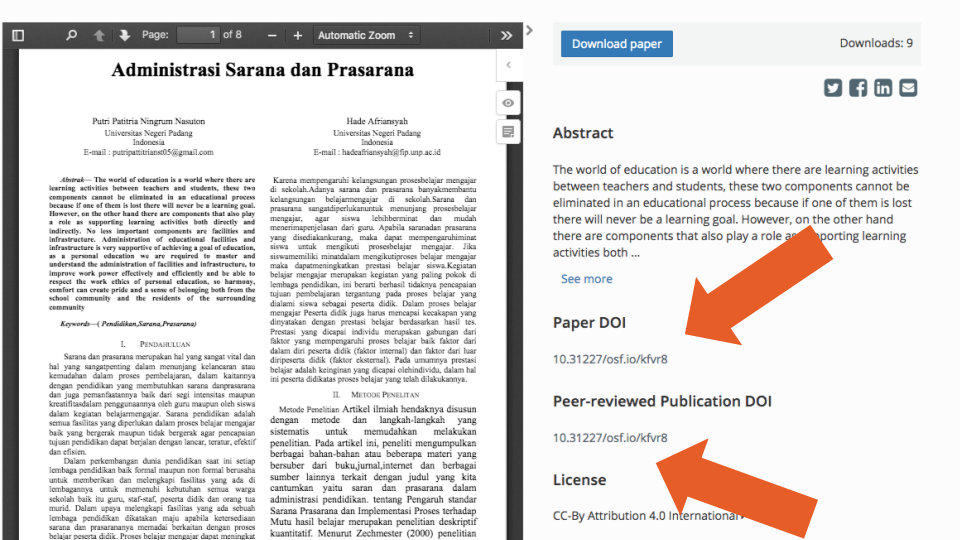
However, after exploring the data, it became apparent that this field alone cannot be used as a clear indicator of whether or not the record corresponds to a preprint (e.g., example one, where the peer-reviewed publication DOI points to an identical PDF that was uploaded as a preprint). In 491 (10%) of cases, the “peer reviewed” DOI pointed to another OSF preprint (e.g., example two, where the DOI points to a duplicate of the same record). Furthermore, 70 (14%) of the records used the record’s own DOI as the peer-reviewed publication DOI (e.g., example three, Figure 7 below). We also identified cases where the DOI entered could not be resolved (i.e., link does not work, as in example four). In addition, we found 362 (7%) cases where DOIs pointed to preprints or data deposited at the Zenodo repository and 9 (0.2%) cases were they pointed to items deposited at the Figshare repository. Finally, we found 305 (6%) records that shared the same Peer-reviewed publication DOI with at least one other record in an OSF repository. In many cases, this was due to the preprints being duplicates of each other, but in others, the error seemed to be a data entry issue (e.g., example five: records one and two are different preprints that point to the same published paper).
Proceed with caution
As can be seen from above, using OSF metadata to answer even the most basic questions about preprints growth and uptake is problematic. The contributor metadata, in particular, was very unreliable, making it difficult to conduct any analyses that relied on author information, including searching for possible postprints using the authors names. We therefore caution researchers and stakeholders citing the number of preprints as reported on the OSF website or in recently published scientific articles1,2,3. To the best of our knowledge, no analyses conducted so far have fully addressed the issues we encountered.
Note: We communicated many of the issues described above with the OSF team at the Centre for Open Science who have indicated they are taking steps to address them. The most recent version of the OSF roadmap is available here.
References
- Narock T, Goldstein EB. Quantifying the Growth of Preprint Services Hosted by the Center for Open Science. Publications. 2019;7(2):44.
- Rahim R, Irawan DE, Zulfikar A, Hardi R, Gultom E, Ginting G, et al., editors. INA-Rxiv: The Missing Puzzle in Indonesia’s Scientific Publishing Workflow. Journal of Physics: Conference Series; 2018: IOP Publishing.
- Riegelman A. OSF Preprints. The Charleston Advisor. 2018;19(3):35-8.
Comments? Questions? Drop us a line on Twitter (tag #scholcommlab). We have shared the issues outlined above with the COS team, and welcome your insights and experiences with working with preprint metadata. Our source code is available on GitHub.
Analyzing preprints: The challenges of working with SHARE metadata
By Mario Malički, Maria Janina Sarol, and Juan Pablo Alperin
As can be seen from a series of recent publications,1-6 there is great interest surrounding preprints—scholarly manuscripts that are published ahead of peer review.

And while these studies show a rise in the number of documents posted on preprint servers, they fail to offer a clear picture on how the growing number of preprint servers are being used, and how different communities are engaging with preprints. That’s why ASAPbio teamed up with the Scholarly Communications Lab on a project investigating preprint uptake and use across life and other sciences.
As we will describe in this four-part blog series, we encountered many challenges in working with preprint metadata, including lack of documentation, missing values, and incompatible and erroneous data. We have documented these challenges, in the hopes of helping those, who like ourselves, would like to better understand what is happening with preprints.
We begin this series with an analysis of the resource that was supposed to aggregate metadata of multiple preprint sources—the SHARE database—while the other three blogs will focus on the Open Science Framework (OSF) preprint servers, bioRxiv, and arXiv.
A deep dive into the Center for Open Science’s SHARE database
The Center for Open Science (COS)—the same group behind the OSF Preprint servers—partnered with the Association of Research Libraries to build the SHARE Infrastructure in order to harvest and normalize metadata from more than 100 sources (preprint servers). Within the 2,222,495 records contained in SHARE as of January 2019 (full dataset is available here, 1.46 GB uncompressed, provided to us by the SHARE team), we uncovered many discrepancies, omissions, and errors, which ultimately prevented us in using it as a source for our planned analyses. We present or findings for each of the major metadata fields available in SHARE:
1) Sources and records per source
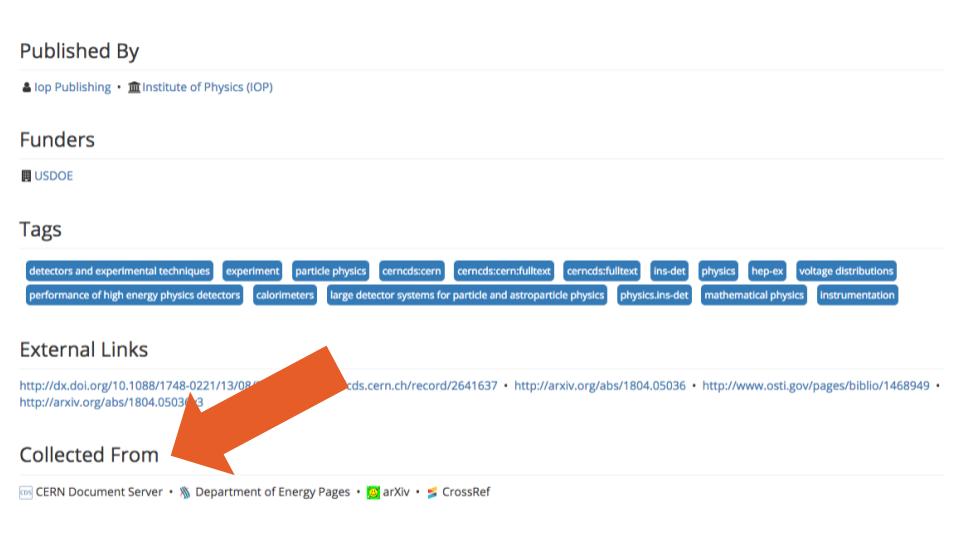
We could not find information on SHARE’s website or GitHub pages specifying to what extent records are harvested from the different sources available in SHARE. In the records database they provided us, we identified 103 unique sources (servers). But we also found some that did not contain all of records that were available at the sources themselves (e.g., there were only 24 preprints from SSRN, out of the more than 700,000 preprints housed there today). In addition, although SHARE attempts to consolidate information from multiple sources for each record (e.g., see Figure 1, below, for a record with information available from four sources), we also found instances where information from different records was consolidated as if it all corresponded to the same record (e.g., example, where information on two different arXiv records—records one and two—was merged, most likely due to the fact that both of these records link to the same postprint publication). A clear description on how information is harvested from each source could help researchers attempting to use this data.
2) Subjects (i.e. scientific discipline or subdiscipline classification).
We were also unable to find documentation on how SHARE handles subject/discipline classifications from various sources, nor on whether there were plans to use a single classification system for all records. We did confirm that, of the records that had a subject field value, 98% used the bepress classification of disciplines. However, this was only 1 of 21 different classifications we identified in the database.
Most records though, 1,517,874 (68%), did not have subject information at all. We felt more could have been done to fill in the subject classifications for these records. For example, records from the RePEc preprint server—which deals exclusively with Economics—could be augmented under the subject Social and Behavioural Sciences, as per their bepress classification, and similarly records from the various arXiv repositories could be augmented based on their respective fields.
3) Dates
Dates are a complicated metadata field, as research documents can be time stamped at many different moments: when they are uploaded, when their DOI was minted, when they were made public, updated, retracted, etc. However, the SHARE metadata contains only two date fields for each record that are populated using the source’s metadata (an additional three date fields exist for timestamps that occur within the SHARE database itself). These two source-based dates are: date_published (defined as: “date when the record was first published, issued, or made publicly available in any form”) and date_updated (defined as: “date when the record was last updated in the source”).
However, 2,085,123 (94%) of the records we examined didn’t have a date_published in SHARE, and only had the date_updated. This made it impossible to distinguish whether the date_updated referred to the date that the record was first created, when its DOI was minted, or when it was published in a printed or online version of a journal. In other words, we couldn’t use this data to determine the elapsed time between a preprint and its published paper (postprint), or to confirm whether the record was a preprint at all (we’ll expand on this issue of preprint/postprint identification in our next blog post). For researchers and users who rely on this database, a clear indication of which dates are harvested from each source would be greatly welcome.
4) Contributors
In the SHARE database, a Contributor is categorized as either a Creator (defined as: “the agent who authored or was a major contributor to the creation of the work”) or as a Contributor (defined as: “the people or organizations responsible for making contributions to an object”). In more scholarly familiar terms, Creators are authors and Contributors are uploaders, i.e. those who uploaded initial or revised versions of the preprints. Any Creator (author) or Contributor (uploader) is further classified as being either a person, institution, organization, or consortium.
This classification system poses several challenges. First, because any preprint record can have multiple Creators and multiple Contributors, SHARE metadata cannot be used to determine who initially uploaded the preprint (and filled the information on the authors, keywords, and other preprint details). Instead, the metadata only offers information about who can upload new versions. This, however, is a common issue in most bibliographic databases; information on who initially submitted a manuscript to a journal is seldom provided.
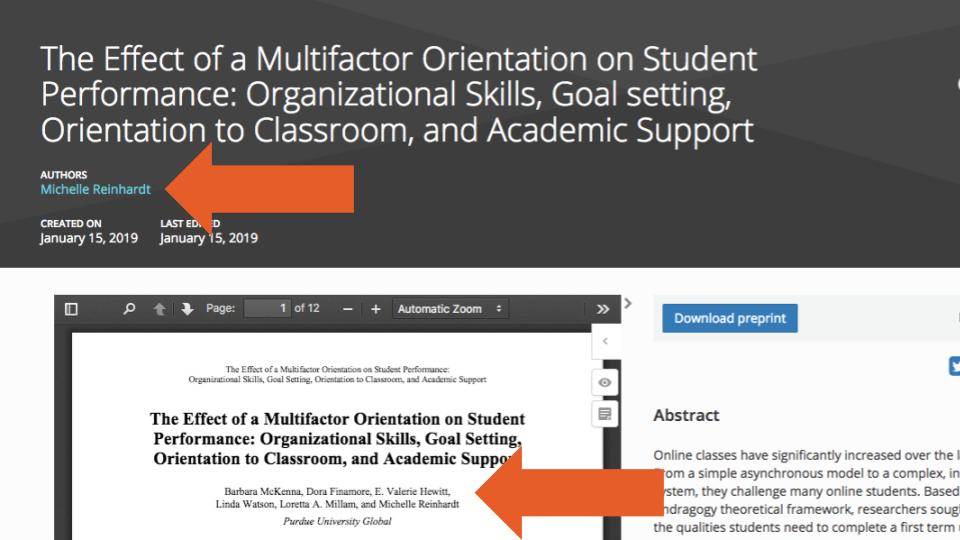
Second, information on the Contributors (uploaders) was available for only 86,579 (4%) of records (most of which were preprints hosted of the OSF preprint servers, arXiv or HAL). While it could be assumed that the Contributors (uploaders) would most often also be Creators (authors) of those records, we found—using a simple exact name matching—that this occurred in only in 2,737 (3%) of the 86,579 records. And just as we were initially confused by the two terms, we also found examples where users uploading preprints might have misunderstood them as well. Such is the case shown in Figure 2, where one person is listed as a Creator (author) and five are listed as Contributors (uploaders), when in fact all six are authors.
Affiliation information was almost non-existent for either authors or uploaders. We found no affiliation information for 2,153,579 (97%) of records.
Even more problematic were records where the number of authors in the metadata did not match the number of authors in the uploaded documents (e.g., this example lists one person as the only Creator and Contributor of the record, but the uploaded PDF has two additional authors, see below; and this example lists one Creator in the metadata, but three authors in the PDF—none of whom is the supposed Creator).
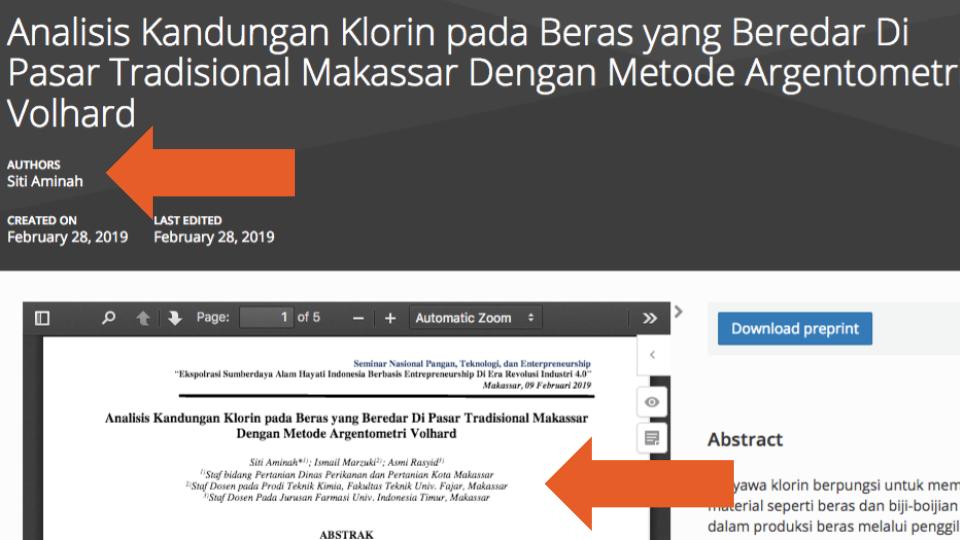
These errors make it very difficult to answer even the most basic preprint authorship questions reliably: How many authors does a specific preprint have? And what is the average (or median) number of authors per preprint? These issues also constrain more detailed investigations into authorship networks; author affiliations, countries or regions of origin, seniority; and identifying all preprints published by a single author.
“These errors make it very difficult to answer even the most basic preprint authorship questions reliably: How many authors does a specific preprint have? And what is the average number of authors per preprint?”
Comparing the metadata of each record with the information in the uploaded documents seems to be the only way to detect all of these errors. Therefore, we can only presume (and hope) that the percentage of records with an incorrect number of authors is small and will not distort our analyses. Preprint servers, however, should apply better methods or checks to ensure information on authors is correctly entered into the metadata when preprints are submitted or posted.
Unfortunately, these were not the only issues that would make analysis of preprint authorship challenging. In SHARE, the byline order position (i.e. place in the list of authors) is provided in the metadata field order_cited for each author or Creator. We found that 454,866 (20%) of records assigned the same byline order position to several different Creators (authors). To the best of our knowledge, these cases do not represent instances where authors contributed equally to a paper or preprint. Instead, they appear to be metadata errors.
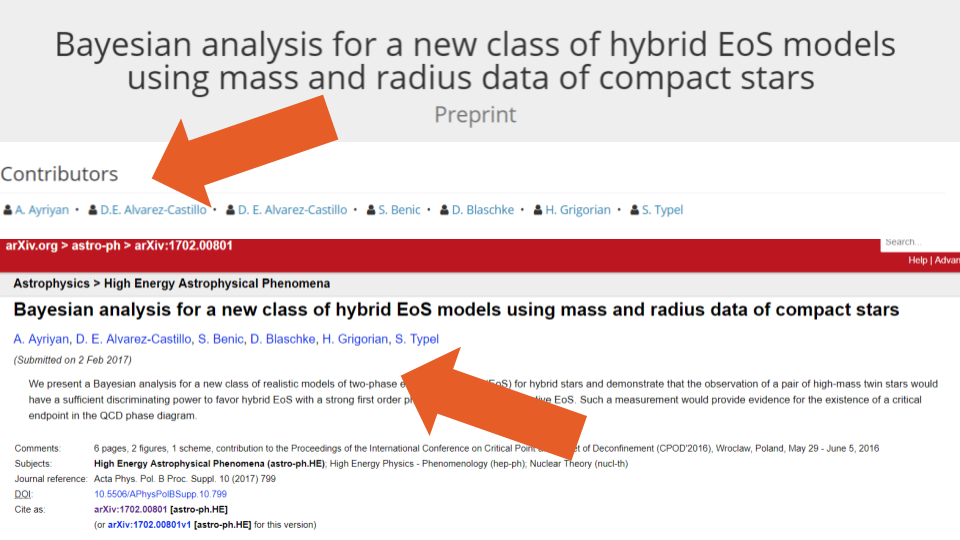
One such error can be found for this preprint, where the metadata lists 4 authors—3 with the same byline order position—while the source’s website shows only two authors and the uploaded PDF only one. Other errors include listing all of the Creators (authors) twice in the metadata (see Figure 4, where 4 authors are included in the uploaded document and 8 in the metadata) and duplicating just one of the names (see Figure 5, which lists 6 authors on the paper, but 7 in the metadata).

We presume these errors most likely occur when the record information is harvested from two different sources, and each source contains a slight variation in the spelling of authors’ names or initials. Additionally, even though we could not find documentation behind the order_cited for Contributors (uploaders), we found examples, as in case of this preprint, where the metadata listed the same byline order position for two uploaders.
To complicate things even further, we also found cases where the byline order listed in the metadata did not match the order in the posted PDF document (e.g., Figure 6 and Figure 7):
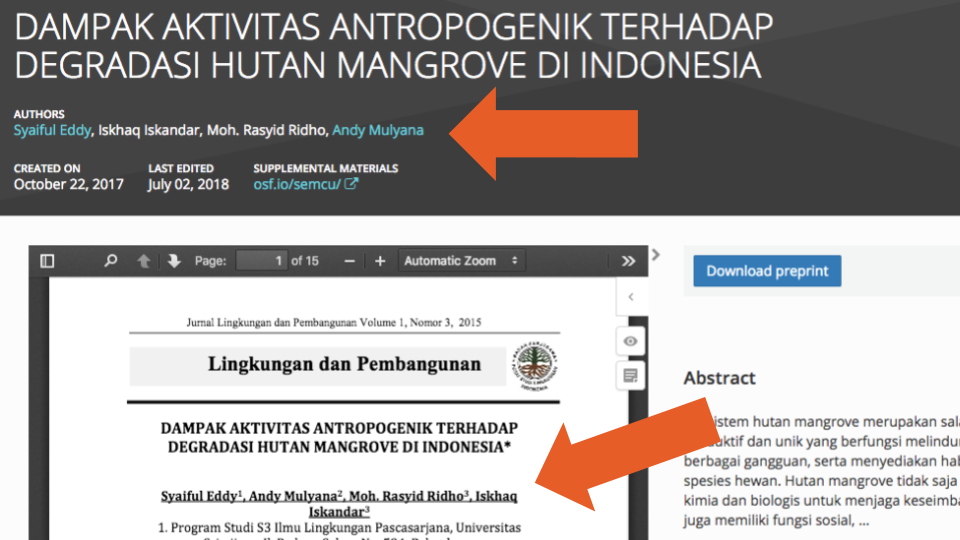
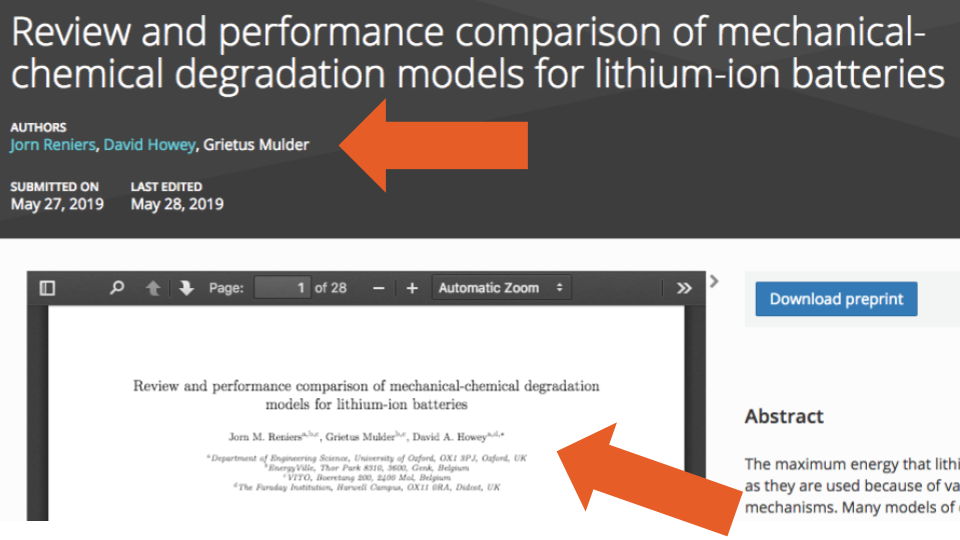
All these Contributor errors we detected mean that if researchers rely solely on the SHARE metadata, any authorship analyses they conduct will likely be very unreliable. We feel more work is needed to improve the quality of authorship information during a creation of a centralized database.
Still a long way to go
As can be seen from the many examples we describe above, even the most basic metadata in SHARE is too problematic to provide a reliable analysis on preprint uptake and use across life or other sciences. The issues we encountered are further compounded by the lack of clear and complete documentation about metadata fields, harvested sources, and the way records from those sources are integrated into the SHARE database. While we cannot attest to the value of SHARE as a service for preprint discovery, we find that, in its current form, it cannot be considered a valuable source for any rigorous analysis of preprints.
In the end, we decided to discard the SHARE database as a starting point for our analyses, and we turned toward the original servers that host that information. Stay tuned for the next post in our series, in which we’ll explore metadata from 24 OSF Preprint servers.
Acknowledgements: We would like to thank Naomi Penfold, Associate Director of ASAPbio, for comments on an earlier version of this post.
Note: We shared a draft of our blog post with the SHARE team at the Centre for Open Science prior to its publication, but did not receive a response at the time of publishing.
References
- Fu DY, Hughey JJ. Releasing a preprint is associated with more attention and citations. bioRxiv. 2019:699652.
- Chiarelli A, Johnson R, Pinfield S, Richens E. Preprints and Scholarly Communication: Adoption, Practices, Drivers and Barriers [version 1; peer review: 2 approved with reservations]. F1000Research. 2019;8(971).
- Fraser N, Momeni F, Mayr P, Peters I. The effect of bioRxiv preprints on citations and altmetrics. bioRxiv. 2019:673665.
- Serghiou S, Ioannidis JP. Altmetric Scores, Citations, and Publication of Studies Posted as Preprints. JAMA: The Journal of the American Medical Association. 2018;319(4):402-4.
- Narock T, Goldstein EB. Quantifying the Growth of Preprint Services Hosted by the Center for Open Science. Publications. 2019;7(2):44.
- Abdill RJ, Blekhman R. Meta-Research: Tracking the popularity and outcomes of all bioRxiv preprints. eLife. 2019;8:e45133.
Comments? Questions? Drop us a line on Twitter (tag #scholcommlab). We have shared the issues outlined above with the COS team, and welcome your insights and experiences with working with preprint metadata. Our source code is available on GitHub.





